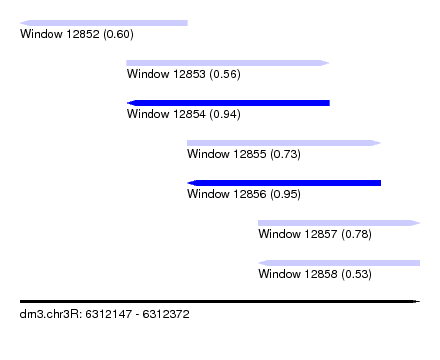
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,312,147 – 6,312,372 |
| Length | 225 |
| Max. P | 0.947665 |

| Location | 6,312,147 – 6,312,241 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 79.56 |
| Shannon entropy | 0.36113 |
| G+C content | 0.60386 |
| Mean single sequence MFE | -26.89 |
| Consensus MFE | -15.84 |
| Energy contribution | -17.40 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.595208 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
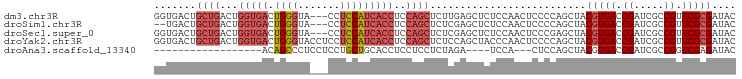
>dm3.chr3R 6312147 94 - 27905053 GGUGACUGCUGACUGGUGACUGGGUA---CCUCCAUCACCUCCAGCUCUUGAGCUCUCCAACUCCCCAGCUACGUGACCGAUCGCCGUUCGCGAUAC (((.((.((((...(((((.((((..---..)))))))))...((((....))))...........))))...)).))).(((((.....))))).. ( -30.60, z-score = -2.12, R) >droSim1.chr3R 12441395 92 + 27517382 --UGACUGCUGACUGGUGACUGGGUA---CCUCCAUCACCUCCAGCUCUCGAGCUCUCCAACUCCCCAGCUACGUGACCGAUCGCCGUUCGCGAUAC --.((..((((...(((((.((((..---..)))))))))..))))..)).((((............)))).(((((.((.....)).))))).... ( -27.20, z-score = -1.72, R) >droSec1.super_0 5491295 94 + 21120651 GGUGACUGCUGACUGGUGACUGGGUA---CCUCCAUCACCUCCAGCUCUCGAGCUCUCCAACUCCCGAGCUACGUGACCGAUCGCCGUUCGCGAUAC (((.((.((((...(((((.((((..---..)))))))))..)))).....(((((..........)))))..)).))).(((((.....))))).. ( -33.50, z-score = -2.48, R) >droYak2.chr3R 10339129 97 - 28832112 GGUGACUGCUGACUGGUGACUGGGUACCUCCUCCAUCACCUCCAGCUCUCCAGCUACCCAACUCCCCAGCUACGUGACCGAUCGCCGUUCGCGAUAC (((.((.((((...(((((.((((.......)))))))))...((((....))))...........))))...)).))).(((((.....))))).. ( -29.80, z-score = -2.04, R) >droAna3.scaffold_13340 22910856 72 + 23697760 ------------------ACAGCCCUCCUCCUGCUGCACCUCCUCCUCUAGA----UCCA---CUCCAGCUACGUGACCGAUCGCCGGUCGAGAUAC ------------------.........(((..((((.......((.....))----....---...)))).....(((((.....)))))))).... ( -13.34, z-score = -0.21, R) >consensus GGUGACUGCUGACUGGUGACUGGGUA___CCUCCAUCACCUCCAGCUCUCGAGCUCUCCAACUCCCCAGCUACGUGACCGAUCGCCGUUCGCGAUAC .......((((...(((((.((((.......)))))))))..))))..........................(((((.((.....)).))))).... (-15.84 = -17.40 + 1.56)
| Location | 6,312,207 – 6,312,321 |
|---|---|
| Length | 114 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 74.20 |
| Shannon entropy | 0.51808 |
| G+C content | 0.57109 |
| Mean single sequence MFE | -37.30 |
| Consensus MFE | -19.60 |
| Energy contribution | -20.21 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.564021 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
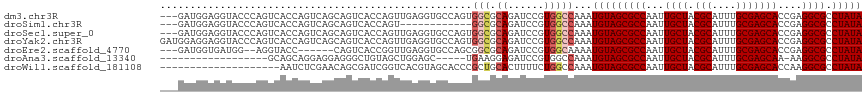
>dm3.chr3R 6312207 114 + 27905053 ---GAUGGAGGUACCCAGUCACCAGUCAGCAGUCACCAGUUGAGGUGCCAGUGGCGCAGAUCCGUGGCCAAAUGUAGCGCCAAUUGCUACGCAUUUGCGAGCACCGAGGCGCCUAUA ---(((((......))).)).....(((((........)))))((((((.(((.(((((((.((((((.((.((......)).)))))))).)))))))..)))...)))))).... ( -39.10, z-score = -0.85, R) >droSim1.chr3R 12441455 102 - 27517382 ---GAUGGAGGUACCCAGUCACCAGUCAGCAGUCACCAGU------------GGCGCAGAUCCGUGGCCAAAUGUAGCGCCAAUUGCUACGCAUUUGCGAGCACCGAGGCGCCUAUA ---..(((......)))(((((..(((.((.((((....)------------))))).)))..)))))....(((((((((...((((.(((....)))))))....)))).))))) ( -35.40, z-score = -1.42, R) >droSec1.super_0 5491355 114 - 21120651 ---GAUGGAGGUACCCAGUCACCAGUCAGCAGUCACCAGUUGAGGUGCCAGUGGCGCAGAUCCGUGGCCAAAUGUAGCGCCAAUUGCUACGCAUUUGCGAGCACCGAGGCGCCUAUA ---(((((......))).)).....(((((........)))))((((((.(((.(((((((.((((((.((.((......)).)))))))).)))))))..)))...)))))).... ( -39.10, z-score = -0.85, R) >droYak2.chr3R 10339189 117 + 28832112 GAUGGAGGAGGUACCCAGUCACCAGUCAGCAGUCACCAGUUGAGGUGCCAGUGGCGCAGAUCCGUGGCCAAAUGUAGCGCCAAUUGCUACGCAUUUGCGAGCACCGAGGCGCCUAUA (((((.(......)))).)).....(((((........)))))((((((.(((.(((((((.((((((.((.((......)).)))))))).)))))))..)))...)))))).... ( -39.50, z-score = -0.58, R) >droEre2.scaffold_4770 2445865 106 - 17746568 ---GAUGGUGAUGG--AGGUACC------CAGUCACCGGUUGAGGUGCCAGCGGCGCAGAUCCGUGGCAAAAUGUAGCGCCAAUUGCUACGCAUUUGCGAGCACCGAGGCGCCUAUA ---..(((((((((--......)------)).))))))....(((((((..((((((((((.((((((((...(.....)...)))))))).)))))))....))).)))))))... ( -45.20, z-score = -2.46, R) >droAna3.scaffold_13340 22910909 93 - 23697760 ------------------GCAGCAGGAGGAGGGCUGUAGCUGGAGC-----UGAAGGAGAUCCGUGGCCAAAUGUAGCGCCAAUUGCUACGCAUUUGCGAGCAA-AAGGCGCCUAUA ------------------(((...((.(((...((.((((....))-----))..))...)))....))...))).(((((..(((((.(((....))))))))-..)))))..... ( -32.70, z-score = -0.40, R) >droWil1.scaffold_181108 92502 97 + 4707319 --------------------AAUCUCGAACAGCGAUCGGUCACGUAGCACCCGCUGCACUUUUCUGGCCAAAUGUAGCGCCAAUUGCUACGCAUUUGCGAGCACCAAGGCGCCUAUA --------------------....(((.....)))..(((((.(((((....))))).......)))))...(((((((((...((((.(((....)))))))....)))).))))) ( -30.10, z-score = -1.24, R) >consensus ___GAUGGAGGUACCCAGUCACCAGUCAGCAGUCACCAGUUGAGGUGCCAGUGGCGCAGAUCCGUGGCCAAAUGUAGCGCCAAUUGCUACGCAUUUGCGAGCACCGAGGCGCCUAUA ....................................................(((.((......)))))...(((((((((...((((.(((....)))))))....)))).))))) (-19.60 = -20.21 + 0.61)

| Location | 6,312,207 – 6,312,321 |
|---|---|
| Length | 114 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 74.20 |
| Shannon entropy | 0.51808 |
| G+C content | 0.57109 |
| Mean single sequence MFE | -38.14 |
| Consensus MFE | -25.56 |
| Energy contribution | -26.32 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.939632 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
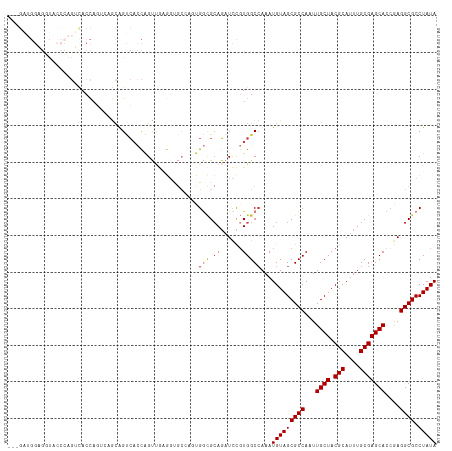
>dm3.chr3R 6312207 114 - 27905053 UAUAGGCGCCUCGGUGCUCGCAAAUGCGUAGCAAUUGGCGCUACAUUUGGCCACGGAUCUGCGCCACUGGCACCUCAACUGGUGACUGCUGACUGGUGACUGGGUACCUCCAUC--- ...(((..((.((((((((((....))).)))...((((((...(((((....)))))..))))))....(((((((.(.(....).).)))..))))))))))..))).....--- ( -41.00, z-score = -1.07, R) >droSim1.chr3R 12441455 102 + 27517382 UAUAGGCGCCUCGGUGCUCGCAAAUGCGUAGCAAUUGGCGCUACAUUUGGCCACGGAUCUGCGCC------------ACUGGUGACUGCUGACUGGUGACUGGGUACCUCCAUC--- ...(((..((.((((((((((....))).)))....(((((...(((((....)))))..)))))------------((..((........))..)).))))))..))).....--- ( -36.20, z-score = -0.69, R) >droSec1.super_0 5491355 114 + 21120651 UAUAGGCGCCUCGGUGCUCGCAAAUGCGUAGCAAUUGGCGCUACAUUUGGCCACGGAUCUGCGCCACUGGCACCUCAACUGGUGACUGCUGACUGGUGACUGGGUACCUCCAUC--- ...(((..((.((((((((((....))).)))...((((((...(((((....)))))..))))))....(((((((.(.(....).).)))..))))))))))..))).....--- ( -41.00, z-score = -1.07, R) >droYak2.chr3R 10339189 117 - 28832112 UAUAGGCGCCUCGGUGCUCGCAAAUGCGUAGCAAUUGGCGCUACAUUUGGCCACGGAUCUGCGCCACUGGCACCUCAACUGGUGACUGCUGACUGGUGACUGGGUACCUCCUCCAUC ...(((..((.((((((((((....))).)))...((((((...(((((....)))))..))))))....(((((((.(.(....).).)))..))))))))))..)))........ ( -41.00, z-score = -0.93, R) >droEre2.scaffold_4770 2445865 106 + 17746568 UAUAGGCGCCUCGGUGCUCGCAAAUGCGUAGCAAUUGGCGCUACAUUUUGCCACGGAUCUGCGCCGCUGGCACCUCAACCGGUGACUG------GGUACCU--CCAUCACCAUC--- ....((((((....(((((((....))).))))...))))))......((((((((.......))).)))))........(((((.((------((....)--))))))))...--- ( -43.20, z-score = -3.01, R) >droAna3.scaffold_13340 22910909 93 + 23697760 UAUAGGCGCCUU-UUGCUCGCAAAUGCGUAGCAAUUGGCGCUACAUUUGGCCACGGAUCUCCUUCA-----GCUCCAGCUACAGCCCUCCUCCUGCUGC------------------ ....((((((..-((((((((....))).)))))..)))))).....((((...(((.((.....)-----).))).))))((((.........)))).------------------ ( -32.20, z-score = -2.35, R) >droWil1.scaffold_181108 92502 97 - 4707319 UAUAGGCGCCUUGGUGCUCGCAAAUGCGUAGCAAUUGGCGCUACAUUUGGCCAGAAAAGUGCAGCGGGUGCUACGUGACCGAUCGCUGUUCGAGAUU-------------------- ....((((((....(((((((....))).))))...))))))................(.(((((((((........)))...)))))).)......-------------------- ( -32.40, z-score = 0.03, R) >consensus UAUAGGCGCCUCGGUGCUCGCAAAUGCGUAGCAAUUGGCGCUACAUUUGGCCACGGAUCUGCGCCACUGGCACCUCAACUGGUGACUGCUGACUGGUGACUGGGUACCUCCAUC___ ....((((((....(((((((....))).))))...)))))).......(((((((.....(((((.............)))))....)))..)))).................... (-25.56 = -26.32 + 0.76)

| Location | 6,312,241 – 6,312,350 |
|---|---|
| Length | 109 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.87 |
| Shannon entropy | 0.31598 |
| G+C content | 0.55179 |
| Mean single sequence MFE | -37.42 |
| Consensus MFE | -27.48 |
| Energy contribution | -27.57 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.729858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
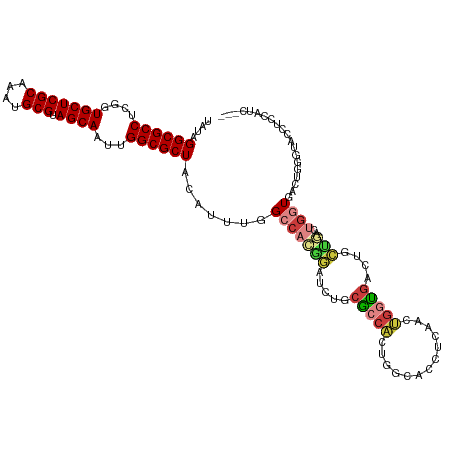
>dm3.chr3R 6312241 109 + 27905053 AGUUGAGGUGCCAGUGGCGCAGAUCCGUGGCCAAAUGUAGCGCCAAUUGCUACGCAUUUGCGAGCACCGAGGCGCCUAUAACAAAUGCCAAUCGUGUCGU-----GGACGAC-GC----- .(((((((((((.(((.(((((((.((((((.((.((......)).)))))))).)))))))..)))...))))))).))))...........((((((.-----...))))-))----- ( -43.90, z-score = -2.19, R) >droSim1.chr3R 12441487 99 - 27517382 ----------CCAGUGGCGCAGAUCCGUGGCCAAAUGUAGCGCCAAUUGCUACGCAUUUGCGAGCACCGAGGCGCCUAUAACAAAUGCCAAUCGUGUCGU-----GGACGAC-GC----- ----------(((.((((((.(((...((((....(((((((((...((((.(((....)))))))....)))).)))))......))))))))))))))-----)).....-..----- ( -35.20, z-score = -1.04, R) >droSec1.super_0 5491389 109 - 21120651 AGUUGAGGUGCCAGUGGCGCAGAUCCGUGGCCAAAUGUAGCGCCAAUUGCUACGCAUUUGCGAGCACCGAGGCGCCUAUAACAAAUGCCAAUCGUGUCGU-----GGACGAC-GC----- .(((((((((((.(((.(((((((.((((((.((.((......)).)))))))).)))))))..)))...))))))).))))...........((((((.-----...))))-))----- ( -43.90, z-score = -2.19, R) >droYak2.chr3R 10339226 114 + 28832112 AGUUGAGGUGCCAGUGGCGCAGAUCCGUGGCCAAAUGUAGCGCCAAUUGCUACGCAUUUGCGAGCACCGAGGCGCCUAUAACAAAUGCCAAUCGUGUCGUUGUGUGGACGAC-GC----- .(((((((((((.(((.(((((((.((((((.((.((......)).)))))))).)))))))..)))...))))))).))))...........((((((((.....))))))-))----- ( -45.80, z-score = -2.12, R) >droEre2.scaffold_4770 2445891 109 - 17746568 GGUUGAGGUGCCAGCGGCGCAGAUCCGUGGCAAAAUGUAGCGCCAAUUGCUACGCAUUUGCGAGCACCGAGGCGCCUAUAACAAAUGCCAAUCGUGUCGU-----GGACGAC-GC----- .(((((((((((..((((((((((.((((((((...(.....)...)))))))).)))))))....))).))))))).))))...........((((((.-----...))))-))----- ( -44.60, z-score = -1.95, R) >droAna3.scaffold_13340 22910928 102 - 23697760 -----AGCUGGAGCUGAAGGAGAUCCGUGGCCAAAUGUAGCGCCAAUUGCUACGCAUUUGCGAGCAA-AAGGCGCCUAUAACAAAUGCCAAUCGUGUCGU-----GG-CGAC-GC----- -----(((....)))((.((....)).((((....(((((((((..(((((.(((....))))))))-..)))).)))))......)))).))((((((.-----..-))))-))----- ( -35.80, z-score = -1.11, R) >dp4.chr2 23639262 92 - 30794189 -----------------UGCAGCUCCGUGGCCAAAUGUAGCGCCAAUUGCUACGCAUUUGCGAGCACCAAGGCGCCUAUAACAAAUGCCAAUCGUGUCGU-----GGACGAC-GC----- -----------------..........((((....(((((((((...((((.(((....)))))))....)))).)))))......))))...((((((.-----...))))-))----- ( -31.90, z-score = -1.17, R) >droPer1.super_0 10812706 92 - 11822988 -----------------UGCAGCUCCGUGGCCAAAUGUAGCGCCAAUUGCUACGCAUUUGCGAGCACCAAGGCGCCUAUAACAAAUGCCAAUCGUGUCGU-----GGACGAC-GC----- -----------------..........((((....(((((((((...((((.(((....)))))))....)))).)))))......))))...((((((.-----...))))-))----- ( -31.90, z-score = -1.17, R) >droWil1.scaffold_181108 92519 109 + 4707319 GGUCACGUAGCACCCGCUGCACUUUUCUGGCCAAAUGUAGCGCCAAUUGCUACGCAUUUGCGAGCACCAAGGCGCCUAUAACAAAUGCCAAUCGUGUCGU-----GGACGAC-GC----- ......(((((....))))).......((((....(((((((((...((((.(((....)))))))....)))).)))))......))))...((((((.-----...))))-))----- ( -38.70, z-score = -1.84, R) >droVir3.scaffold_13047 1277886 91 + 19223366 -----------------UGCA-UUUUGUGGCCAAAUGUAGCGCCAAUUGCUACGCAUUUGCGAGCACCCAGGCGCCUAUAACAAAUGCCAAUCGUGUCGU-----GGACGAC-GC----- -----------------....-.....((((....(((((((((...((((.(((....)))))))....)))).)))))......))))...((((((.-----...))))-))----- ( -31.90, z-score = -1.62, R) >droMoj3.scaffold_6540 5343921 91 + 34148556 -----------------UGCA-UUUUGUGGCCAAAUGUAGCGCCAAUUGCUACGCAUUUGCGAGCACCGAGGCGCCUAUAACAAAUGCCAAUCGUGUCGU-----GGACGAC-GC----- -----------------....-.....((((....(((((((((...((((.(((....)))))))....)))).)))))......))))...((((((.-----...))))-))----- ( -31.90, z-score = -1.44, R) >droGri2.scaffold_14906 9192229 97 - 14172833 -----------------UGCA-GUUUGUGGCCAAAUGUAGCGCCAAUUGCUACGCAUUUGCGAGCACCAAGGCGCCUAUAACAAAUGCCAAUCGUGUCGU-----GUACGUUUGCGACGC -----------------....-.....((((....(((((((((...((((.(((....)))))))....)))).)))))......))))...(((((((-----(......)))))))) ( -33.50, z-score = -1.04, R) >consensus __________C____G_UGCAGAUCCGUGGCCAAAUGUAGCGCCAAUUGCUACGCAUUUGCGAGCACCGAGGCGCCUAUAACAAAUGCCAAUCGUGUCGU_____GGACGAC_GC_____ ...........................((((....(((((((((...((((.(((....)))))))....)))).)))))......))))...(((((((.......))))).))..... (-27.48 = -27.57 + 0.09)


| Location | 6,312,241 – 6,312,350 |
|---|---|
| Length | 109 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.87 |
| Shannon entropy | 0.31598 |
| G+C content | 0.55179 |
| Mean single sequence MFE | -38.08 |
| Consensus MFE | -32.38 |
| Energy contribution | -32.71 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.947665 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
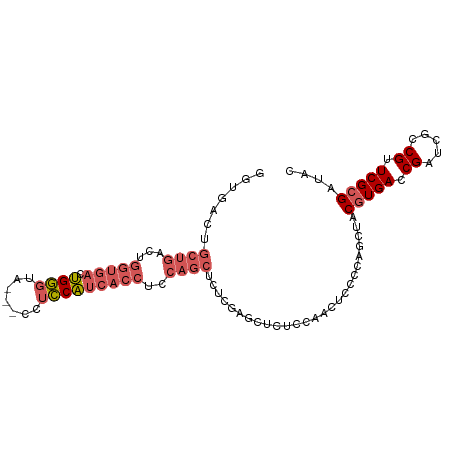
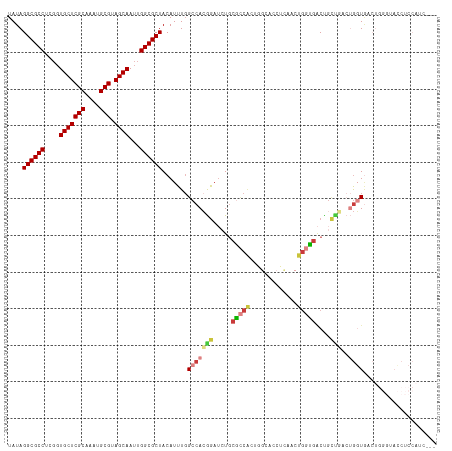
>dm3.chr3R 6312241 109 - 27905053 -----GC-GUCGUCC-----ACGACACGAUUGGCAUUUGUUAUAGGCGCCUCGGUGCUCGCAAAUGCGUAGCAAUUGGCGCUACAUUUGGCCACGGAUCUGCGCCACUGGCACCUCAACU -----((-((((...-----.)))).((..((((...(((....((((((....(((((((....))).))))...)))))))))....)))))).....))(((...)))......... ( -39.50, z-score = -1.40, R) >droSim1.chr3R 12441487 99 + 27517382 -----GC-GUCGUCC-----ACGACACGAUUGGCAUUUGUUAUAGGCGCCUCGGUGCUCGCAAAUGCGUAGCAAUUGGCGCUACAUUUGGCCACGGAUCUGCGCCACUGG---------- -----((-((((...-----.)))).......((((((((....(((((....))))).))))))))...))...((((((...(((((....)))))..))))))....---------- ( -38.00, z-score = -1.54, R) >droSec1.super_0 5491389 109 + 21120651 -----GC-GUCGUCC-----ACGACACGAUUGGCAUUUGUUAUAGGCGCCUCGGUGCUCGCAAAUGCGUAGCAAUUGGCGCUACAUUUGGCCACGGAUCUGCGCCACUGGCACCUCAACU -----((-((((...-----.)))).((..((((...(((....((((((....(((((((....))).))))...)))))))))....)))))).....))(((...)))......... ( -39.50, z-score = -1.40, R) >droYak2.chr3R 10339226 114 - 28832112 -----GC-GUCGUCCACACAACGACACGAUUGGCAUUUGUUAUAGGCGCCUCGGUGCUCGCAAAUGCGUAGCAAUUGGCGCUACAUUUGGCCACGGAUCUGCGCCACUGGCACCUCAACU -----((-(((((.......))))).((..((((...(((....((((((....(((((((....))).))))...)))))))))....)))))).....))(((...)))......... ( -39.70, z-score = -1.41, R) >droEre2.scaffold_4770 2445891 109 + 17746568 -----GC-GUCGUCC-----ACGACACGAUUGGCAUUUGUUAUAGGCGCCUCGGUGCUCGCAAAUGCGUAGCAAUUGGCGCUACAUUUUGCCACGGAUCUGCGCCGCUGGCACCUCAACC -----((-((((...-----.)))).((..(((((..(((....((((((....(((((((....))).))))...)))))))))...))))))).....))(((...)))......... ( -40.90, z-score = -1.63, R) >droAna3.scaffold_13340 22910928 102 + 23697760 -----GC-GUCG-CC-----ACGACACGAUUGGCAUUUGUUAUAGGCGCCUU-UUGCUCGCAAAUGCGUAGCAAUUGGCGCUACAUUUGGCCACGGAUCUCCUUCAGCUCCAGCU----- -----((-((((-..-----.)))).....((((...(((....((((((..-((((((((....))).)))))..)))))))))....)))).(((.((.....)).))).)).----- ( -38.70, z-score = -2.85, R) >dp4.chr2 23639262 92 + 30794189 -----GC-GUCGUCC-----ACGACACGAUUGGCAUUUGUUAUAGGCGCCUUGGUGCUCGCAAAUGCGUAGCAAUUGGCGCUACAUUUGGCCACGGAGCUGCA----------------- -----((-((((...-----.)))).((..((((...(((....((((((....(((((((....))).))))...)))))))))....)))))).....)).----------------- ( -36.50, z-score = -1.70, R) >droPer1.super_0 10812706 92 + 11822988 -----GC-GUCGUCC-----ACGACACGAUUGGCAUUUGUUAUAGGCGCCUUGGUGCUCGCAAAUGCGUAGCAAUUGGCGCUACAUUUGGCCACGGAGCUGCA----------------- -----((-((((...-----.)))).((..((((...(((....((((((....(((((((....))).))))...)))))))))....)))))).....)).----------------- ( -36.50, z-score = -1.70, R) >droWil1.scaffold_181108 92519 109 - 4707319 -----GC-GUCGUCC-----ACGACACGAUUGGCAUUUGUUAUAGGCGCCUUGGUGCUCGCAAAUGCGUAGCAAUUGGCGCUACAUUUGGCCAGAAAAGUGCAGCGGGUGCUACGUGACC -----..-((((...-----.))))(((..(((((((((((...((((((....(((((((....))).))))...)))))).(((((........))))).)))))))))))))).... ( -41.90, z-score = -1.35, R) >droVir3.scaffold_13047 1277886 91 - 19223366 -----GC-GUCGUCC-----ACGACACGAUUGGCAUUUGUUAUAGGCGCCUGGGUGCUCGCAAAUGCGUAGCAAUUGGCGCUACAUUUGGCCACAAAA-UGCA----------------- -----((-((((...-----.)))).....((((...(((....((((((....(((((((....))).))))...)))))))))....)))).....-.)).----------------- ( -35.80, z-score = -2.20, R) >droMoj3.scaffold_6540 5343921 91 - 34148556 -----GC-GUCGUCC-----ACGACACGAUUGGCAUUUGUUAUAGGCGCCUCGGUGCUCGCAAAUGCGUAGCAAUUGGCGCUACAUUUGGCCACAAAA-UGCA----------------- -----((-((((...-----.)))).....((((...(((....((((((....(((((((....))).))))...)))))))))....)))).....-.)).----------------- ( -35.80, z-score = -2.49, R) >droGri2.scaffold_14906 9192229 97 + 14172833 GCGUCGCAAACGUAC-----ACGACACGAUUGGCAUUUGUUAUAGGCGCCUUGGUGCUCGCAAAUGCGUAGCAAUUGGCGCUACAUUUGGCCACAAAC-UGCA----------------- ((((((.........-----.)))).....((((...(((....((((((....(((((((....))).))))...)))))))))....)))).....-.)).----------------- ( -34.10, z-score = -1.35, R) >consensus _____GC_GUCGUCC_____ACGACACGAUUGGCAUUUGUUAUAGGCGCCUCGGUGCUCGCAAAUGCGUAGCAAUUGGCGCUACAUUUGGCCACGGAUCUGCA_C____G__________ .....((.(((((.......))))).....((((...(((....((((((....(((((((....))).))))...)))))))))....)))).......)).................. (-32.38 = -32.71 + 0.33)

| Location | 6,312,281 – 6,312,372 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 92.31 |
| Shannon entropy | 0.15513 |
| G+C content | 0.50105 |
| Mean single sequence MFE | -26.19 |
| Consensus MFE | -25.92 |
| Energy contribution | -25.71 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.08 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.99 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.780809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6312281 91 + 27905053 CGCCAAUUGCUACGCAUUUGCGAGCACCGAGGCGCCUAUAACAAAUGCCAAUCGUGUCGU------GGACGACGCCAGCGAUAUUUAAUUGUUUCUG------------------ ((((...((((.(((....)))))))....))))...................((((((.------...))))))(((.((((......)))).)))------------------ ( -25.40, z-score = -0.80, R) >droSim1.chr3R 12441517 91 - 27517382 CGCCAAUUGCUACGCAUUUGCGAGCACCGAGGCGCCUAUAACAAAUGCCAAUCGUGUCGU------GGACGACGCCAGCGAUAUUUAAUUGUUUCUG------------------ ((((...((((.(((....)))))))....))))...................((((((.------...))))))(((.((((......)))).)))------------------ ( -25.40, z-score = -0.80, R) >droSec1.super_0 5491429 91 - 21120651 CGCCAAUUGCUACGCAUUUGCGAGCACCGAGGCGCCUAUAACAAAUGCCAAUCGUGUCGU------GGACGACGCCAGCGAUAUUUAAUUGUUUCUG------------------ ((((...((((.(((....)))))))....))))...................((((((.------...))))))(((.((((......)))).)))------------------ ( -25.40, z-score = -0.80, R) >droYak2.chr3R 10339266 96 + 28832112 CGCCAAUUGCUACGCAUUUGCGAGCACCGAGGCGCCUAUAACAAAUGCCAAUCGUGUCGUUGUGU-GGACGACGCCAGCGAUAUUUAAUUGUUUCUG------------------ ((((...((((.(((....)))))))....))))..............((((.(((((((((.((-(.....))))))))))))...))))......------------------ ( -29.20, z-score = -1.24, R) >droEre2.scaffold_4770 2445931 91 - 17746568 CGCCAAUUGCUACGCAUUUGCGAGCACCGAGGCGCCUAUAACAAAUGCCAAUCGUGUCGU------GGACGACGCCAGCGAUAUUUAAUUGUUUCUG------------------ ((((...((((.(((....)))))))....))))...................((((((.------...))))))(((.((((......)))).)))------------------ ( -25.40, z-score = -0.80, R) >droAna3.scaffold_13340 22910963 89 - 23697760 CGCCAAUUGCUACGCAUUUGCGAGCAA-AAGGCGCCUAUAACAAAUGCCAAUCGUGUCGUG-------GCGACGCCAGCGAUAUUUAAUUGUUUCUG------------------ ((((..(((((.(((....))))))))-..))))..............((((.((((((((-------((...)))).))))))...))))......------------------ ( -27.60, z-score = -1.55, R) >dp4.chr2 23639285 91 - 30794189 CGCCAAUUGCUACGCAUUUGCGAGCACCAAGGCGCCUAUAACAAAUGCCAAUCGUGUCGU------GGACGACGCCAGCGAUAUUUAAUUGUUUCUG------------------ ((((...((((.(((....)))))))....))))...................((((((.------...))))))(((.((((......)))).)))------------------ ( -25.40, z-score = -1.08, R) >droPer1.super_0 10812729 91 - 11822988 CGCCAAUUGCUACGCAUUUGCGAGCACCAAGGCGCCUAUAACAAAUGCCAAUCGUGUCGUGG------ACGACGCCAGCGAUAUUUAAUUGUUUCUG------------------ ((((...((((.(((....)))))))....))))...................((((((...------.))))))(((.((((......)))).)))------------------ ( -25.40, z-score = -1.08, R) >droWil1.scaffold_181108 92559 109 + 4707319 CGCCAAUUGCUACGCAUUUGCGAGCACCAAGGCGCCUAUAACAAAUGCCAAUCGUGUCGU------GGACGACGCCAGCGAUAUUUAAUUGUUUUUGCCGCCACCACCACCACCA .......((((.(((....)))))))....((((.(...(((((.((...(((((((((.------...))))....)))))...)).)))))...).))))............. ( -27.30, z-score = -0.72, R) >droVir3.scaffold_13047 1277908 91 + 19223366 CGCCAAUUGCUACGCAUUUGCGAGCACCCAGGCGCCUAUAACAAAUGCCAAUCGUGUCGU------GGACGACGCCAGCGAUAUUUAAUUGUUUCUG------------------ ((((...((((.(((....)))))))....))))...................((((((.------...))))))(((.((((......)))).)))------------------ ( -25.40, z-score = -1.14, R) >droMoj3.scaffold_6540 5343943 91 + 34148556 CGCCAAUUGCUACGCAUUUGCGAGCACCGAGGCGCCUAUAACAAAUGCCAAUCGUGUCGU------GGACGACGCCAGCGAUAUUUAAUUGUUUCUG------------------ ((((...((((.(((....)))))))....))))...................((((((.------...))))))(((.((((......)))).)))------------------ ( -25.40, z-score = -0.80, R) >droGri2.scaffold_14906 9192251 97 - 14172833 CGCCAAUUGCUACGCAUUUGCGAGCACCAAGGCGCCUAUAACAAAUGCCAAUCGUGUCGUGUACGUUUGCGACGCCAGCGAUAUUUAAUUGUUUCUG------------------ ((((...((((.(((....)))))))....))))...................((((((((......))))))))(((.((((......)))).)))------------------ ( -27.00, z-score = -0.85, R) >consensus CGCCAAUUGCUACGCAUUUGCGAGCACCGAGGCGCCUAUAACAAAUGCCAAUCGUGUCGU______GGACGACGCCAGCGAUAUUUAAUUGUUUCUG__________________ ((((...((((.(((....)))))))....))))...................(((((((........)))))))(((.((((......)))).))).................. (-25.92 = -25.71 + -0.21)
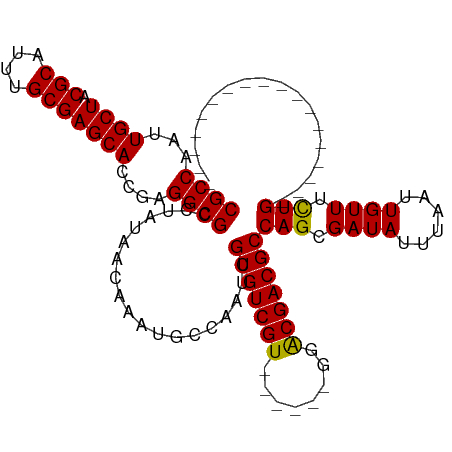
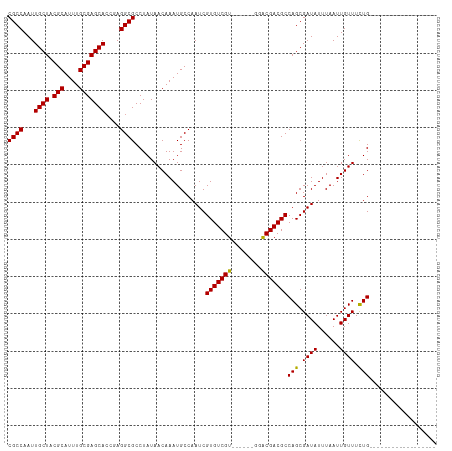
| Location | 6,312,281 – 6,312,372 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 92.31 |
| Shannon entropy | 0.15513 |
| G+C content | 0.50105 |
| Mean single sequence MFE | -28.58 |
| Consensus MFE | -27.05 |
| Energy contribution | -27.30 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.87 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.527853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6312281 91 - 27905053 ------------------CAGAAACAAUUAAAUAUCGCUGGCGUCGUCC------ACGACACGAUUGGCAUUUGUUAUAGGCGCCUCGGUGCUCGCAAAUGCGUAGCAAUUGGCG ------------------.................((((.((((((...------.)))).......((((((((....(((((....))))).))))))))...))....)))) ( -27.80, z-score = -0.90, R) >droSim1.chr3R 12441517 91 + 27517382 ------------------CAGAAACAAUUAAAUAUCGCUGGCGUCGUCC------ACGACACGAUUGGCAUUUGUUAUAGGCGCCUCGGUGCUCGCAAAUGCGUAGCAAUUGGCG ------------------.................((((.((((((...------.)))).......((((((((....(((((....))))).))))))))...))....)))) ( -27.80, z-score = -0.90, R) >droSec1.super_0 5491429 91 + 21120651 ------------------CAGAAACAAUUAAAUAUCGCUGGCGUCGUCC------ACGACACGAUUGGCAUUUGUUAUAGGCGCCUCGGUGCUCGCAAAUGCGUAGCAAUUGGCG ------------------.................((((.((((((...------.)))).......((((((((....(((((....))))).))))))))...))....)))) ( -27.80, z-score = -0.90, R) >droYak2.chr3R 10339266 96 - 28832112 ------------------CAGAAACAAUUAAAUAUCGCUGGCGUCGUCC-ACACAACGACACGAUUGGCAUUUGUUAUAGGCGCCUCGGUGCUCGCAAAUGCGUAGCAAUUGGCG ------------------.................((((.(((((((..-.....))))).......((((((((....(((((....))))).))))))))...))....)))) ( -28.00, z-score = -0.89, R) >droEre2.scaffold_4770 2445931 91 + 17746568 ------------------CAGAAACAAUUAAAUAUCGCUGGCGUCGUCC------ACGACACGAUUGGCAUUUGUUAUAGGCGCCUCGGUGCUCGCAAAUGCGUAGCAAUUGGCG ------------------.................((((.((((((...------.)))).......((((((((....(((((....))))).))))))))...))....)))) ( -27.80, z-score = -0.90, R) >droAna3.scaffold_13340 22910963 89 + 23697760 ------------------CAGAAACAAUUAAAUAUCGCUGGCGUCGC-------CACGACACGAUUGGCAUUUGUUAUAGGCGCCUU-UUGCUCGCAAAUGCGUAGCAAUUGGCG ------------------((((..(((((.....(((.((((...))-------)))))...)))))...)))).......((((..-((((((((....))).)))))..)))) ( -28.00, z-score = -1.50, R) >dp4.chr2 23639285 91 + 30794189 ------------------CAGAAACAAUUAAAUAUCGCUGGCGUCGUCC------ACGACACGAUUGGCAUUUGUUAUAGGCGCCUUGGUGCUCGCAAAUGCGUAGCAAUUGGCG ------------------.................((((.((((((...------.)))).......((((((((....(((((....))))).))))))))...))....)))) ( -27.80, z-score = -0.92, R) >droPer1.super_0 10812729 91 + 11822988 ------------------CAGAAACAAUUAAAUAUCGCUGGCGUCGU------CCACGACACGAUUGGCAUUUGUUAUAGGCGCCUUGGUGCUCGCAAAUGCGUAGCAAUUGGCG ------------------.................((((.((((((.------...)))).......((((((((....(((((....))))).))))))))...))....)))) ( -27.80, z-score = -0.92, R) >droWil1.scaffold_181108 92559 109 - 4707319 UGGUGGUGGUGGUGGCGGCAAAAACAAUUAAAUAUCGCUGGCGUCGUCC------ACGACACGAUUGGCAUUUGUUAUAGGCGCCUUGGUGCUCGCAAAUGCGUAGCAAUUGGCG ..((.((((((..(.((((.................)))).)..)).))------)).)).((((((((((((((....(((((....))))).))))))))....))))))... ( -38.43, z-score = -1.06, R) >droVir3.scaffold_13047 1277908 91 - 19223366 ------------------CAGAAACAAUUAAAUAUCGCUGGCGUCGUCC------ACGACACGAUUGGCAUUUGUUAUAGGCGCCUGGGUGCUCGCAAAUGCGUAGCAAUUGGCG ------------------.................((((.((((((...------.)))).......((((((((....(((((....))))).))))))))...))....)))) ( -27.80, z-score = -0.75, R) >droMoj3.scaffold_6540 5343943 91 - 34148556 ------------------CAGAAACAAUUAAAUAUCGCUGGCGUCGUCC------ACGACACGAUUGGCAUUUGUUAUAGGCGCCUCGGUGCUCGCAAAUGCGUAGCAAUUGGCG ------------------.................((((.((((((...------.)))).......((((((((....(((((....))))).))))))))...))....)))) ( -27.80, z-score = -0.90, R) >droGri2.scaffold_14906 9192251 97 + 14172833 ------------------CAGAAACAAUUAAAUAUCGCUGGCGUCGCAAACGUACACGACACGAUUGGCAUUUGUUAUAGGCGCCUUGGUGCUCGCAAAUGCGUAGCAAUUGGCG ------------------.................((((.((((((..........)))).......((((((((....(((((....))))).))))))))...))....)))) ( -26.10, z-score = 0.16, R) >consensus __________________CAGAAACAAUUAAAUAUCGCUGGCGUCGUCC______ACGACACGAUUGGCAUUUGUUAUAGGCGCCUCGGUGCUCGCAAAUGCGUAGCAAUUGGCG ...................................((((.(((((((........))))).......((((((((....(((((....))))).))))))))...))....)))) (-27.05 = -27.30 + 0.25)
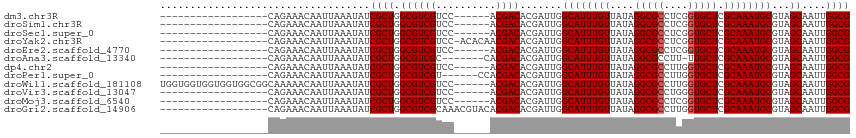
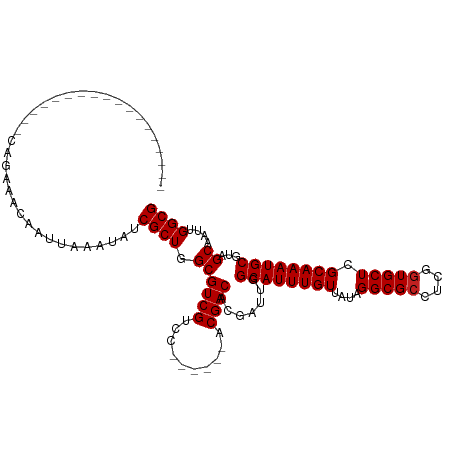
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:03:15 2011