| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,308,728 – 6,308,820 |
| Length | 92 |
| Max. P | 0.557456 |

| Location | 6,308,728 – 6,308,820 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 71.05 |
| Shannon entropy | 0.56229 |
| G+C content | 0.49096 |
| Mean single sequence MFE | -20.22 |
| Consensus MFE | -7.57 |
| Energy contribution | -7.31 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.60 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.557456 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
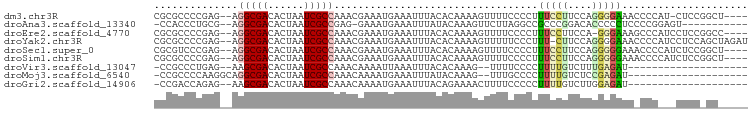
>dm3.chr3R 6308728 92 - 27905053 CGCGCCCCGAG--AGGCGACACUAAUCGCCAAACGAAAUGAAAUUUACACAAAAGUUUUCCCCUUUCCUUCCAGGGGAAACCCCAU-CUCCGGCU---- ...(((..(((--((((((......)))))..........................((((((((........)))))))).....)-))).))).---- ( -27.60, z-score = -3.15, R) >droAna3.scaffold_13340 22907919 84 + 23697760 -CCACCCUGCG--AGGCGACACUAAUCGCCGAG-GAAAUGAAAUUUAUACAAAGUUCUUAGGCCGCCCGGACACCCCCUCCCCGGAGU----------- -.......(((--.(((((......)))))(((-((..((.........))...)))))....)))((((...........))))...----------- ( -17.70, z-score = 0.43, R) >droEre2.scaffold_4770 2442641 92 + 17746568 CGCGCCCCGAG--AGGCGACACUAAUCGCCAAACGAAAUGAAAUUUACACAAAAGUUUUCCCCUUUCCUUCCA-GGGAAAGCCCAUCCUCCGGCC---- ...(((..(((--.(((((......)))))........................((((((((...........-)))))))).....))).))).---- ( -24.00, z-score = -2.23, R) >droYak2.chr3R 10335648 96 - 28832112 CGCGCCCCGAG--AGGCGACACUAAUCGCCAAACGAAAUGAAAUUUACACAAAAGUUUUUCCCUUU-CUUCCAGGGGAAACCCCAUCCUCCAGCUAGAU ...((...(((--.(((((......)))))....((((.(((((((......)))))))....)))-).....(((....)))....)))..))..... ( -21.80, z-score = -1.68, R) >droSec1.super_0 5487944 93 + 21120651 CGCGUCCCGAG--AGGCGACACUAAUCGCCAAACGAAAUGAAAUUUACACAAAAGUUUUCCCCUUUCCUUCCAGGGGGAAACCCCAUCUCCGGCU---- ...(.((.(((--((((((......)))))........................((((((((((........))))))))))....)))).))).---- ( -29.30, z-score = -2.95, R) >droSim1.chr3R 12438071 93 + 27517382 CGCGCCCCGAG--AGGCGACACUAAUCGCCAAACGAAAUGAAAUUUACACAAAAGUUUUCCCCUUUCCUUCCAGGGGGAAACCCCAUCUCCGGCU---- ...(((..(((--((((((......)))))........................((((((((((........))))))))))....)))).))).---- ( -31.00, z-score = -3.59, R) >droVir3.scaffold_13047 1274530 74 - 19223366 -CCGCCCUGAG--AAGCGACACUAAUCGCCAAACAAAAUUAAAUUUACACAAAG--UUUUCCCCUUUUGUCUUUGAGAU-------------------- -.....((.((--(.((((......))))...((((((..((((((.....)))--))).....)))))).))).))..-------------------- ( -7.60, z-score = -0.44, R) >droMoj3.scaffold_6540 5339445 76 - 34148556 -CCGCCCCAAGGCAGGCGACACUAAUCGCCAAACAAAAUGAAAUUUAUACAAAG--UUUGCCCCUUUUGUCUCCGAGAU-------------------- -..(((....))).(((((......)))))..((((((.(((((((.....)))--)))...).)))))).........-------------------- ( -13.90, z-score = -1.15, R) >droGri2.scaffold_14906 9187696 76 + 14172833 -CCGACCAGAG--AAGCGACACUAAUCGCCAAACAAAAUGAAAUUUACAGAAAACUUUUCCCCCUUUUGUCUUGGAGAU-------------------- -(((((.((((--..((((......))))..........((((.............))))...)))).)))..))....-------------------- ( -9.12, z-score = -0.26, R) >consensus CGCGCCCCGAG__AGGCGACACUAAUCGCCAAACGAAAUGAAAUUUACACAAAAGUUUUCCCCUUUCCUUCCAGGGGAAA_CCC___CUCC_GC_____ ..............(((((......)))))..................................(((((....)))))..................... ( -7.57 = -7.31 + -0.26)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:03:08 2011