| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,279,296 – 6,279,441 |
| Length | 145 |
| Max. P | 0.896726 |

| Location | 6,279,296 – 6,279,441 |
|---|---|
| Length | 145 |
| Sequences | 7 |
| Columns | 158 |
| Reading direction | forward |
| Mean pairwise identity | 75.11 |
| Shannon entropy | 0.48815 |
| G+C content | 0.47878 |
| Mean single sequence MFE | -43.74 |
| Consensus MFE | -26.17 |
| Energy contribution | -27.32 |
| Covariance contribution | 1.15 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.896726 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
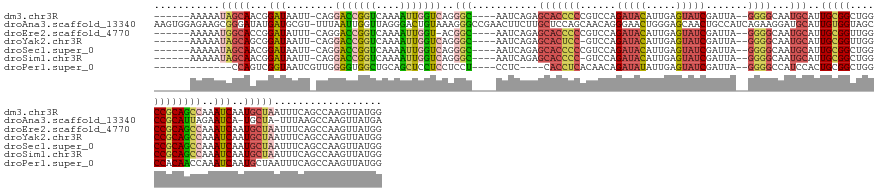
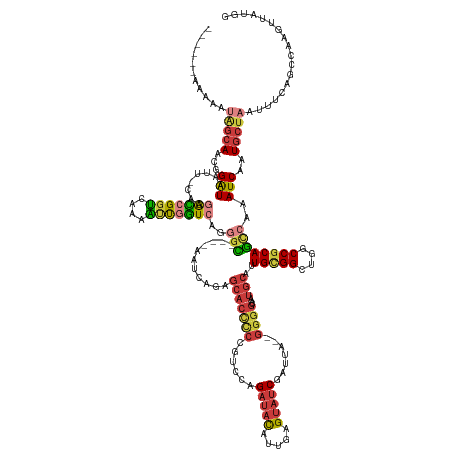
>dm3.chr3R 6279296 145 + 27905053 ------AAAAAUAGCAACGGAUAAUU-CAGGACCGGUCAAAAUUGGUCAGGGC----AAUCAGAGCACCCCCGUCCAGAUACAUUGAGUAUCGAUUA--GGGGCAAUGCAUUGCGGCUGGCCGCAGCCAAAUCAAUGCUAAUUUCAGCCAAGUUAUGG ------.....(((((...(((....-...(((((((....)))))))..(((----.......(((.((((.....(((((.....))))).....--))))...)))..(((((....))))))))..)))..))))).......(((.....))) ( -45.50, z-score = -2.14, R) >droAna3.scaffold_13340 22880115 155 - 23697760 AAGUGGAGAAGCGGGAUAUGAUGCGU-UUUAAUUGGUUAGGGACUGUAAAGGGCCGAACUUCUUGCUCCAGCAACAGGGAACUGGGAGCAACUGCCAUCAGAAGGAUGCAUUGUGGUAGCCCGCAUUAGAAUCA-UGCUA-UUUAAGCCAAGUUAUGA ...(((....(((((.((..((((((-(((...((((..((..((....))..)).......(((((((.....(((....))))))))))..)))).....)))))))))..))....)))))..(((.....-..)))-......)))........ ( -45.40, z-score = -0.38, R) >droEre2.scaffold_4770 2411242 144 - 17746568 ------AAAAAUGGCACCGGAUAUUU-CAGGACCGGUCAAAAUUGGU-ACGGC----AAUCAGAGCACCCCCGUCCAGAUACAUUGAGUAUCGAUUA--GGGGCAAUGCAUUGCGGUUGGCCGCAGCCAAAUCAAUGCUAAUUUCAGCCAAGUUAUGG ------.....(((((((((......-.....)))))....((((((-..(((----.......(((.((((.....(((((.....))))).....--))))...)))..(((((....))))))))..))))))..........))))........ ( -46.20, z-score = -1.98, R) >droYak2.chr3R 10309433 144 + 28832112 ------AAAAAUAGCAGCGGAUAAUU-CAGGACCGGUCAAAAUUGGUCAGGGC----AAUCAGAGCACUCC-GUCCAGAUACAUUGAGUAUCGAUUA--GGGGCAAUGCAUUGCGGUUGGCCGCAGCCAAAUCAAUGCUAAUUUCAGCCAAGUUAUGG ------.....(((((...(((....-...(((((((....)))))))..(((----.......(((((((-.....(((((.....))))).....--))))...)))..(((((....))))))))..)))..))))).......(((.....))) ( -43.10, z-score = -1.97, R) >droSec1.super_0 5458780 145 - 21120651 ------AAAAAUAGCAACGGAUAAUU-CAGGACCGGUCAAAAUUGGUCAGGGC----AAUCAGAGCACCCCCGUCCAGAUACAUUGAGUAUCGAUUA--GGGGCAAUGCAUUGCGGCUGGCCGCAGCCAAAUCAAUGCUAAUUUCAGCCAAGUUAUGG ------.....(((((...(((....-...(((((((....)))))))..(((----.......(((.((((.....(((((.....))))).....--))))...)))..(((((....))))))))..)))..))))).......(((.....))) ( -45.50, z-score = -2.14, R) >droSim1.chr3R 12406189 144 - 27517382 ------AAAAAUAGCAACGGAUAAUU-CAGGACCGGUCAAAAUUGGUCAGGGC----AAUCAGAGCACCCC-GUCCAGAUACAUUGAGUAUCGAUUA--GGGGCAAUGCAUUGCGGCUGGCCGCAGCCAAAUCAAUGCUAAUUUCAGCCAAGUUAUGG ------.....(((((...(((....-...(((((((....)))))))..(((----.......(((((((-.....(((((.....))))).....--))))...)))..(((((....))))))))..)))..))))).......(((.....))) ( -45.20, z-score = -2.26, R) >droPer1.super_0 10779677 135 - 11822988 -------------CCAGUCGGUAAUCGUUGGGGUGGCUGCAGCUCCUCCUCCU----CCUC----CACCUCACAACAGAUAUAUUGAGUAUCGAUUA--GGGGCCAUCCACUGCGGCUGGCCACAACCAAAUCAAUGCUAAUUUCAGCCAAGUUAUGG -------------(((.(((((((((((((((((((..(.((......)).).----...)----)))).).)))).))).))))))((((.((((.--(((((((.((.....)).)))))....)).)))).)))).................))) ( -35.30, z-score = 0.42, R) >consensus ______AAAAAUAGCAACGGAUAAUU_CAGGACCGGUCAAAAUUGGUCAGGGC____AAUCAGAGCACCCCCGUCCAGAUACAUUGAGUAUCGAUUA__GGGGCAAUGCAUUGCGGCUGGCCGCAGCCAAAUCAAUGCUAAUUUCAGCCAAGUUAUGG ...........(((((...(((........(((((((....)))))))..(((...........(((((((......(((((.....))))).......))))...)))..(((((....))))))))..)))..))))).................. (-26.17 = -27.32 + 1.15)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:03:02 2011