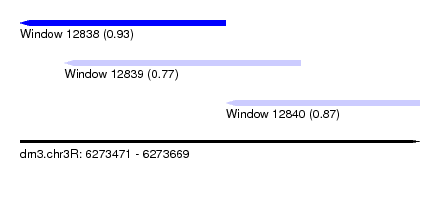
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,273,471 – 6,273,669 |
| Length | 198 |
| Max. P | 0.932020 |

| Location | 6,273,471 – 6,273,573 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 87.37 |
| Shannon entropy | 0.27186 |
| G+C content | 0.44500 |
| Mean single sequence MFE | -25.68 |
| Consensus MFE | -20.04 |
| Energy contribution | -19.97 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.932020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6273471 102 - 27905053 AGCCGCCGAAAAACGAUAAAUUCAAUCGAAAAUCGUUACAUGUCAAUGCAUUAAUCGGCCAUAAUCCCAUUAUGGCGCAAGGAUAUG--CCCUUGCUGUCCCUG----------- .((((((((..((((((...(((....))).))))))..((((....))))...))))).(((((...))))))))((((((.....--.))))))........----------- ( -27.00, z-score = -2.62, R) >droEre2.scaffold_4770 2405540 102 + 17746568 AGCCGCCGAAAAACGAUAAAUUCAAUCGAAAAUCGUUACAUGUCAAUGCAUUAAUCGGCCAUAAUCCCAUUAUGGCGCAAGGAUAUG--CCCUUGCUGUCCCUG----------- .((((((((..((((((...(((....))).))))))..((((....))))...))))).(((((...))))))))((((((.....--.))))))........----------- ( -27.00, z-score = -2.62, R) >droYak2.chr3R 10303458 103 - 28832112 AGCCGCCGAAAAACGAUAAAUUCAAUCGAAAAUCGUUACAUGUCAAUGCAUUAAUCGGCCAUAAUCCCAUUAUGGCGCAAGGAUAUG-CCCCUUGCUGUCCCUG----------- .((((((((..((((((...(((....))).))))))..((((....))))...))))).(((((...))))))))((((((.....-..))))))........----------- ( -26.50, z-score = -2.55, R) >droSec1.super_0 5453024 102 + 21120651 AGCCGCCGAAAAACGAUAAAUUCAAUCGAAAAUCGUUACAUGUCAAUGCAUUAAUCGGCCAUAAUCCCAUUAUGGCGCAAGGAUAUG--CCCUUGCUGUCCCUG----------- .((((((((..((((((...(((....))).))))))..((((....))))...))))).(((((...))))))))((((((.....--.))))))........----------- ( -27.00, z-score = -2.62, R) >droSim1.chr3R 12400403 102 + 27517382 AGCCGCCGAAAAACGAUAAAUUCAAUCGAAAAUCGUUACAUGUCAAUGCAUUAAUCGGCCAUAAUCCCAUUAUGGCGCAAGGAUAUG--CCCUUGCUGUCCCUG----------- .((((((((..((((((...(((....))).))))))..((((....))))...))))).(((((...))))))))((((((.....--.))))))........----------- ( -27.00, z-score = -2.62, R) >dp4.chr2 23600081 108 + 30794189 AGCCGCCGAAAAACGAUAAAUUCAAUCGAAAAUCGUUACAUGUCAAUGCAUUAAUCGGCCAUAAUCCCAUUAUGGCGCAAGGAUAUGCC-CCGCCAAGUCCUCUCCCUC------ .((((((((..((((((...(((....))).))))))..((((....))))...))))).(((((...))))))))...(((((.((..-....)).))))).......------ ( -22.40, z-score = -1.43, R) >droPer1.super_0 10773545 108 + 11822988 AGCCGCCGAAAAACGAUAAAUUCAAUCGAAAAUCGUUACAUGUCAAUGCAUUAAUCGGCCAUAAUCCCAUUAUGGCGCAAGGAUAUGCC-CCGCCAAGUCCUCUCCCUC------ .((((((((..((((((...(((....))).))))))..((((....))))...))))).(((((...))))))))...(((((.((..-....)).))))).......------ ( -22.40, z-score = -1.43, R) >droAna3.scaffold_13340 22875428 105 + 23697760 AGCCGCCGAAAAACGAUAAAUUCAAUCGAAAAUCGUUACAUGUCAAUGCAUUAAUCGGCCAUAAUCCCAUUAUGGCGCAAGGAUGUGUGUCCUCUCCGUCCCACA---------- .((((((((..((((((...(((....))).))))))..((((....))))...))))).(((((...))))))))....(((((.(.(....).))))))....---------- ( -22.80, z-score = -1.09, R) >droWil1.scaffold_181108 50957 114 - 4707319 AGCCGCCAAAAAACGAUAAAUUCAAUCGAAAAUCGUUACAUGUCAAUGCAUUAAUCGGCCAUAAUCCCAUUAUGGCGCAAGGAUAUGCCACCAGCAUGUCCCCUCUCUCAAUUG- ....((.....((((((...(((....))).))))))....................((((((((...))))))))))..((((((((.....)))))))).............- ( -26.30, z-score = -2.72, R) >droVir3.scaffold_13047 1235963 111 - 19223366 AGCCGCCGAAAAACGAUAAAUUCAAUCGAAAAUCGUUACAUGUCAAUGCAUUAAUCGGCCAUAAUCCCAUUAUGGCGCAAGGAUAUG---CCGGC-ACUCCUCUGAAUGUCUCUC ....((((...((((((...(((....))).)))))).((((((..(((........((((((((...)))))))))))..))))))---.))))-................... ( -28.20, z-score = -2.13, R) >droMoj3.scaffold_6540 5281728 112 - 34148556 AGCCGCCGAAAAACGAUAAAUUCAAUCGAAAAUCGUUACAUGUCAAUGCAUUAAUCGGCCAUAAUCCCAUUAUGGCGCAAGGAUAUG---CCGAAAAGUCCUCUCAAUGUCCCUC .((((((((..((((((...(((....))).))))))..((((....))))...))))).(((((...))))))))....((((((.---..((.......))...))))))... ( -23.20, z-score = -1.40, R) >droGri2.scaffold_14906 9147025 112 + 14172833 AGCCGCCGAAAAACGAUAAAUUCAAUCGAAAAUCGUUACAUGUCAAUGCAUUAAUCGGCCAUAAUCCCAUUAUGGCGCAAGGAUAUG---CCGGCAAGUCCUCUGAACGUCGCUC (((.((((...((((((...(((....))).)))))).((((((..(((........((((((((...)))))))))))..))))))---.)))).((....)).......))). ( -28.30, z-score = -1.45, R) >consensus AGCCGCCGAAAAACGAUAAAUUCAAUCGAAAAUCGUUACAUGUCAAUGCAUUAAUCGGCCAUAAUCCCAUUAUGGCGCAAGGAUAUG___CCUUCCUGUCCCCU___________ ...........((((((...(((....))).)))))).((((((..(((........((((((((...)))))))))))..))))))............................ (-20.04 = -19.97 + -0.08)
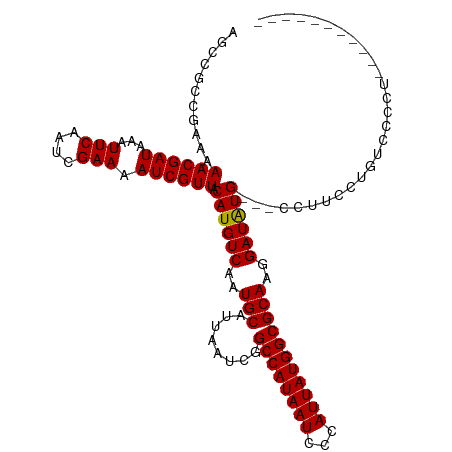
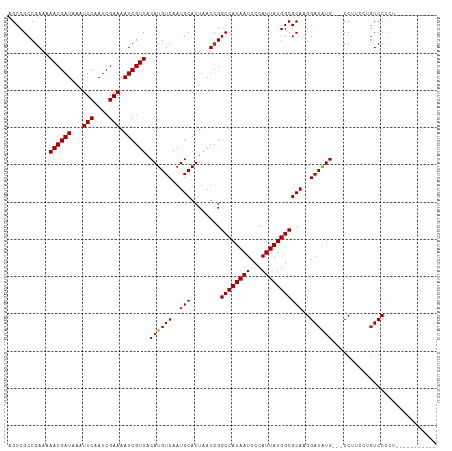
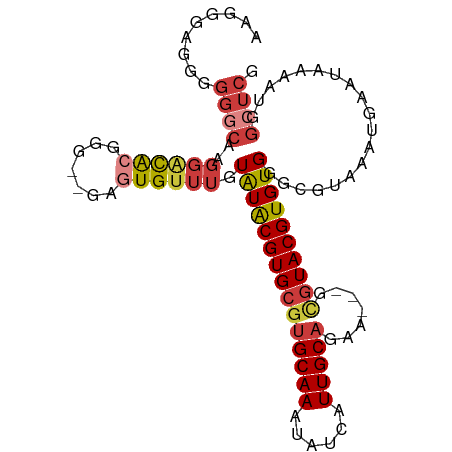
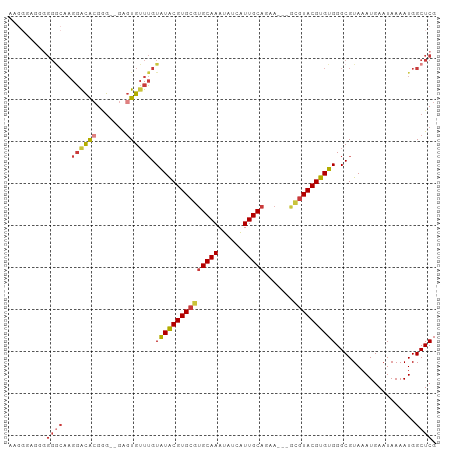
| Location | 6,273,493 – 6,273,610 |
|---|---|
| Length | 117 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.48 |
| Shannon entropy | 0.05456 |
| G+C content | 0.41578 |
| Mean single sequence MFE | -30.87 |
| Consensus MFE | -29.68 |
| Energy contribution | -29.37 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.96 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.768047 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6273493 117 - 27905053 ---AAGCGUACGUGUGGGCGUAAAUGAAUAAAAUGGCUCGAGCCGCCGAAAAACGAUAAAUUCAAUCGAAAAUCGUUACAUGUCAAUGCAUUAAUCGGCCAUAAUCCCAUUAUGGCGCAA ---..((((....(((((.....(((........(((....)))(((((..((((((...(((....))).))))))..((((....))))...))))))))...)))))....)))).. ( -32.10, z-score = -1.56, R) >droEre2.scaffold_4770 2405562 117 + 17746568 ---AAGCGUACGUGUGGGCGUAAAUGAAUAAAAUGGCUCGAGCCGCCGAAAAACGAUAAAUUCAAUCGAAAAUCGUUACAUGUCAAUGCAUUAAUCGGCCAUAAUCCCAUUAUGGCGCAA ---..((((....(((((.....(((........(((....)))(((((..((((((...(((....))).))))))..((((....))))...))))))))...)))))....)))).. ( -32.10, z-score = -1.56, R) >droYak2.chr3R 10303481 117 - 28832112 ---AAGCGUACGUGUGGGCGUAAAUGAAUAAAAUGGCUCGAGCCGCCGAAAAACGAUAAAUUCAAUCGAAAAUCGUUACAUGUCAAUGCAUUAAUCGGCCAUAAUCCCAUUAUGGCGCAA ---..((((....(((((.....(((........(((....)))(((((..((((((...(((....))).))))))..((((....))))...))))))))...)))))....)))).. ( -32.10, z-score = -1.56, R) >droSec1.super_0 5453046 117 + 21120651 ---AAGCGUACGUGUGGGCGUAAAUGAAUAAAAUGGCUCGAGCCGCCGAAAAACGAUAAAUUCAAUCGAAAAUCGUUACAUGUCAAUGCAUUAAUCGGCCAUAAUCCCAUUAUGGCGCAA ---..((((....(((((.....(((........(((....)))(((((..((((((...(((....))).))))))..((((....))))...))))))))...)))))....)))).. ( -32.10, z-score = -1.56, R) >droSim1.chr3R 12400425 117 + 27517382 ---AAGCGUACGUGUGGGCGUAAAUGAAUAAAAUGGCUCGAGCCGCCGAAAAACGAUAAAUUCAAUCGAAAAUCGUUACAUGUCAAUGCAUUAAUCGGCCAUAAUCCCAUUAUGGCGCAA ---..((((....(((((.....(((........(((....)))(((((..((((((...(((....))).))))))..((((....))))...))))))))...)))))....)))).. ( -32.10, z-score = -1.56, R) >dp4.chr2 23600109 115 + 30794189 -----GCAUACGUGUGGGCGUAAAUGAAUAAAAUGGCUCGAGCCGCCGAAAAACGAUAAAUUCAAUCGAAAAUCGUUACAUGUCAAUGCAUUAAUCGGCCAUAAUCCCAUUAUGGCGCAA -----((((((((((.(((....((.......))(((....))))))....((((((...(((....))).))))))))))))..))))........((((((((...)))))))).... ( -30.30, z-score = -1.30, R) >droPer1.super_0 10773573 115 + 11822988 -----GCAUACGUGUGGGCGUAAAUGAAUAAAAUGGCUCGAGCCGCCGAAAAACGAUAAAUUCAAUCGAAAAUCGUUACAUGUCAAUGCAUUAAUCGGCCAUAAUCCCAUUAUGGCGCAA -----((((((((((.(((....((.......))(((....))))))....((((((...(((....))).))))))))))))..))))........((((((((...)))))))).... ( -30.30, z-score = -1.30, R) >droAna3.scaffold_13340 22875453 120 + 23697760 AACUCAUGUACGUGUGGGCGUAAAUGAAUAAAAUGGCUCGAGCCGCCGAAAAACGAUAAAUUCAAUCGAAAAUCGUUACAUGUCAAUGCAUUAAUCGGCCAUAAUCCCAUUAUGGCGCAA ...((((.(((((....))))).)))).......(((....)))(((((..((((((...(((....))).))))))..((((....))))...)))((((((((...)))))))))).. ( -29.50, z-score = -1.02, R) >droWil1.scaffold_181108 50991 115 - 4707319 -----GCAUAUGUGUGGGCGUAAAUGAAUAAAAUGGCUCGAGCCGCCAAAAAACGAUAAAUUCAAUCGAAAAUCGUUACAUGUCAAUGCAUUAAUCGGCCAUAAUCCCAUUAUGGCGCAA -----((..((((((.(((((.(((((......((((.......)))).....((((.......))))....)))))..))))).))))))......((((((((...)))))))))).. ( -28.90, z-score = -1.22, R) >droVir3.scaffold_13047 1235994 115 - 19223366 -----GCAUACGUGUGGGCGUAAAUGAAUAAAAUGGCUCGAGCCGCCGAAAAACGAUAAAUUCAAUCGAAAAUCGUUACAUGUCAAUGCAUUAAUCGGCCAUAAUCCCAUUAUGGCGCAA -----((((((((((.(((....((.......))(((....))))))....((((((...(((....))).))))))))))))..))))........((((((((...)))))))).... ( -30.30, z-score = -1.30, R) >droMoj3.scaffold_6540 5281760 115 - 34148556 -----GCAUACGUGUGGGCGUAAAUGAAUAAAAUGGCUCGAGCCGCCGAAAAACGAUAAAUUCAAUCGAAAAUCGUUACAUGUCAAUGCAUUAAUCGGCCAUAAUCCCAUUAUGGCGCAA -----((((((((((.(((....((.......))(((....))))))....((((((...(((....))).))))))))))))..))))........((((((((...)))))))).... ( -30.30, z-score = -1.30, R) >droGri2.scaffold_14906 9147057 115 + 14172833 -----GCAUACGUGUGGGCGUAAAUGAAUAAAAUGGCUCGAGCCGCCGAAAAACGAUAAAUUCAAUCGAAAAUCGUUACAUGUCAAUGCAUUAAUCGGCCAUAAUCCCAUUAUGGCGCAA -----((((((((((.(((....((.......))(((....))))))....((((((...(((....))).))))))))))))..))))........((((((((...)))))))).... ( -30.30, z-score = -1.30, R) >consensus _____GCAUACGUGUGGGCGUAAAUGAAUAAAAUGGCUCGAGCCGCCGAAAAACGAUAAAUUCAAUCGAAAAUCGUUACAUGUCAAUGCAUUAAUCGGCCAUAAUCCCAUUAUGGCGCAA .....((((((((((.(((....((.......))(((....))))))....((((((...(((....))).))))))))))))..))))........((((((((...)))))))).... (-29.68 = -29.37 + -0.32)

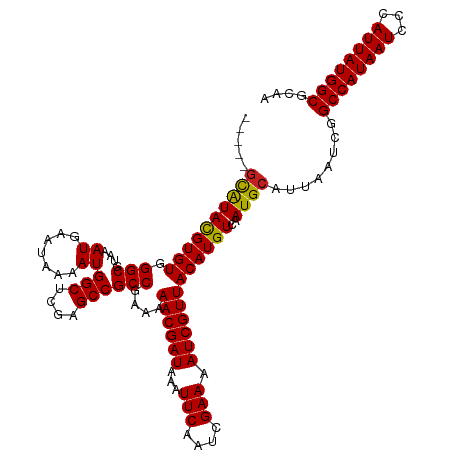
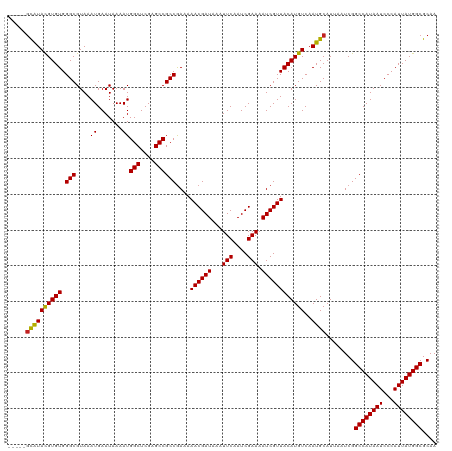
| Location | 6,273,573 – 6,273,669 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 82.50 |
| Shannon entropy | 0.36293 |
| G+C content | 0.50589 |
| Mean single sequence MFE | -26.67 |
| Consensus MFE | -18.30 |
| Energy contribution | -19.37 |
| Covariance contribution | 1.07 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.866158 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6273573 96 - 27905053 GUGGGA-AGGGGCAAGGACACGGG--GAGUGUUUGUAUACGUGCGUGCAAAUAUCAUUGCAGAA---GCGUACGUGUGGGCGUAAAUGAAUAAAAUGGCUCG ......-..((((..((((((...--..)))))).(((((((((((((((......))))....---)))))))))))...................)))). ( -27.50, z-score = -2.22, R) >droEre2.scaffold_4770 2405642 97 + 17746568 AAGCAAGGGGGGCAAGGACACGGG--GAGUGUUUGUAUACGUGCGUGCAAAUAUCAUUGCAGAA---GCGUACGUGUGGGCGUAAAUGAAUAAAAUGGCUCG .........((((..((((((...--..)))))).(((((((((((((((......))))....---)))))))))))...................)))). ( -27.50, z-score = -2.11, R) >droYak2.chr3R 10303561 99 - 28832112 AAAGGGGGGGGGCAAGGACACGGGGAGAGUGUUUGUAUACGUGCGUGCAAAUAUCAUUGCAGAA---GCGUACGUGUGGGCGUAAAUGAAUAAAAUGGCUCG .........((((..((((((.......)))))).(((((((((((((((......))))....---)))))))))))...................)))). ( -27.20, z-score = -2.66, R) >droSec1.super_0 5453126 97 + 21120651 GUGGGAGGGGGGCAAGGACACGGG--GAGUGUUUGUAUACGUGCGUGCAAAUAUCAUUGCAGAA---GCGUACGUGUGGGCGUAAAUGAAUAAAAUGGCUCG .........((((..((((((...--..)))))).(((((((((((((((......))))....---)))))))))))...................)))). ( -27.50, z-score = -2.21, R) >droSim1.chr3R 12400505 97 + 27517382 GAGGGAAGGGGGCAAGGACACGGG--GAGUGUUUGUAUACGUGCGUGCAAAUAUCAUUGCAGAA---GCGUACGUGUGGGCGUAAAUGAAUAAAAUGGCUCG .........((((..((((((...--..)))))).(((((((((((((((......))))....---)))))))))))...................)))). ( -27.50, z-score = -2.64, R) >droAna3.scaffold_13340 22875533 101 + 23697760 ACGGGCCCAGGACGAGGGCAAGGGG-AAGUGUUUGUAUACGUGCGUGCAAAUAUCAUUGCAGAACUCAUGUACGUGUGGGCGUAAAUGAAUAAAAUGGCUCG .((((((..........((((...(-(..(((((((((......))))))))))).)))).....((((.(((((....))))).)))).......)))))) ( -28.40, z-score = -0.72, R) >droVir3.scaffold_13047 1236074 78 - 19223366 ---------------GUGUGCGAAUGGGCUGGUGGUGUGCGUGCGAGCAAAUACCGUUGCA---------UACGUGUGGGCGUAAAUGAAUAAAAUGGCUCG ---------------((((((((......(((((...(((......)))..))))))))))---------)))....((((................)))). ( -21.09, z-score = 0.41, R) >consensus AAGGGAGGGGGGCAAGGACACGGG__GAGUGUUUGUAUACGUGCGUGCAAAUAUCAUUGCAGAA___GCGUACGUGUGGGCGUAAAUGAAUAAAAUGGCUCG .........((((..((((((.......)))))).(((((((((((((((......)))).......)))))))))))...................)))). (-18.30 = -19.37 + 1.07)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:03:00 2011