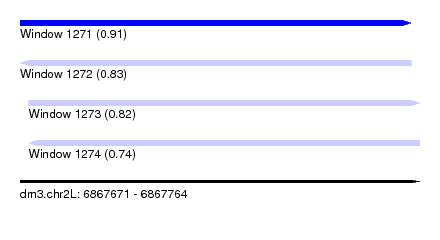
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 6,867,671 – 6,867,764 |
| Length | 93 |
| Max. P | 0.913429 |

| Location | 6,867,671 – 6,867,762 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 60.43 |
| Shannon entropy | 0.68110 |
| G+C content | 0.42211 |
| Mean single sequence MFE | -24.54 |
| Consensus MFE | -9.66 |
| Energy contribution | -11.01 |
| Covariance contribution | 1.35 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.913429 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 6867671 91 + 23011544 UAUUGGACACUUGGCUAAUCUCCACUGGGCUUUUUCCGGCUGC----GAUUAGCCGGUUGUAGGACUGUUAAUUGACUU-UAGCUGGGAAACAUGG-------------------- ......(((.((((((((((.(..(((((.....)))))..).----)))))))))).)))....(.(((((......)-)))).)(....)....-------------------- ( -25.20, z-score = -1.20, R) >droPer1.super_5 4862911 93 + 6813705 -----UAUUUUUGGGCAAU-UCCAUUAGACAAUAUCCGGCAGAAAGGGAAAUUUCAUUUCUAAGAAUGUCAGUUGAUUA-UAGAAAAAAAGG-UAGUGUUG--------------- -----......((((....-))))...((((....((....((((......)))).((((((.....(((....)))..-))))))....))-...)))).--------------- ( -12.50, z-score = 0.78, R) >dp4.chr4_group5 365400 94 + 2436548 -----UAUUUUUGGGCAAA-UCCAUUAGACAAUGUCCGGCAGAAAGGGAAAAUUCAUUUCUAAGAAUGCCAGUUGAUUA-AAGAAAAAAAGGGUAGUGUUG--------------- -----((((((((((((..-((.....))...)))))((((.....(((((.....))))).....)))).........-........)))))))......--------------- ( -16.70, z-score = -0.26, R) >droEre2.scaffold_4929 15786541 85 + 26641161 UAUUGGACACUUGGCUAAUCUCCGCUGGGCUCUUUCCUGCUAC----GAUUAGCCAGUUGUAGGACU--UAGCUAAC----AGCUAAAAUUGUUA--------------------- ......(((.((((((((((...((.(((.....))).))...----)))))))))).))).....(--((((....----.)))))........--------------------- ( -25.00, z-score = -1.93, R) >droYak2.chr2L 16287957 111 - 22324452 UAUUGGACACUUGGCUAAUCUCCGCUGGGCUUUUUCCGGCUAC----GAUUAGCCAGUUGUAGGACUGCUAAUUGACUU-UGGCUAUGACGAUUGCGUUGAGAAUCCAAUUAUUUA .((((((..(((((((((((...((((((.....))))))...----))))))))((((((......(((((......)-))))....)))))).....)))..))))))...... ( -35.50, z-score = -3.47, R) >droSec1.super_3 2407785 79 + 7220098 UAUUGGACACUUGGCUAAUCUCCGCUGGGCUUUUUCCGGCUGC----GAUUAGCCGGUUGUAGGACUGUUAAUUGAAUCAUAG--------------------------------- ......(((.((((((((((.(.((((((.....)))))).).----)))))))))).)))......................--------------------------------- ( -25.50, z-score = -2.42, R) >droSim1.chr2L 6668570 91 + 22036055 UAUUGGACACUUGGCUAAUCUCCGCUGGGCUUUUUCCGGCUGC----GAUUAGCCAGUUGUAGGACUGUUAAUUGCUUU-UAGCUGGGAUUCAUGG-------------------- ...((((..((..(((((.....((((((.....)))))).((----(((((((.((((....)))))))))))))..)-))))..)).))))...-------------------- ( -31.40, z-score = -2.65, R) >consensus UAUUGGACACUUGGCUAAUCUCCGCUGGGCUUUUUCCGGCUGC____GAUUAGCCAGUUGUAGGACUGUUAAUUGACUU_UAGCUAAAAAGG_UAG____________________ ......(((.((((((((((....(((((.....)))))........)))))))))).)))....................................................... ( -9.66 = -11.01 + 1.35)

| Location | 6,867,671 – 6,867,762 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 60.43 |
| Shannon entropy | 0.68110 |
| G+C content | 0.42211 |
| Mean single sequence MFE | -19.46 |
| Consensus MFE | -6.58 |
| Energy contribution | -8.03 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.831587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 6867671 91 - 23011544 --------------------CCAUGUUUCCCAGCUA-AAGUCAAUUAACAGUCCUACAACCGGCUAAUC----GCAGCCGGAAAAAGCCCAGUGGAGAUUAGCCAAGUGUCCAAUA --------------------...((((.....((..-..)).....)))).....(((...((((((((----...((.((.......)).))...))))))))...)))...... ( -18.70, z-score = -1.35, R) >droPer1.super_5 4862911 93 - 6813705 ---------------CAACACUA-CCUUUUUUUCUA-UAAUCAACUGACAUUCUUAGAAAUGAAAUUUCCCUUUCUGCCGGAUAUUGUCUAAUGGA-AUUGCCCAAAAAUA----- ---------------........-............-.........((((.(((((((((.(.......).))))))..)))...))))...(((.-.....)))......----- ( -9.80, z-score = -0.15, R) >dp4.chr4_group5 365400 94 - 2436548 ---------------CAACACUACCCUUUUUUUCUU-UAAUCAACUGGCAUUCUUAGAAAUGAAUUUUCCCUUUCUGCCGGACAUUGUCUAAUGGA-UUUGCCCAAAAAUA----- ---------------............(((((....-.((((..((((((......((((.....))))......)))))).((((....))))))-)).....)))))..----- ( -12.90, z-score = -0.69, R) >droEre2.scaffold_4929 15786541 85 - 26641161 ---------------------UAACAAUUUUAGCU----GUUAGCUA--AGUCCUACAACUGGCUAAUC----GUAGCAGGAAAGAGCCCAGCGGAGAUUAGCCAAGUGUCCAAUA ---------------------.......((((((.----....))))--))....(((..(((((((((----...((.((.......)).))...)))))))))..)))...... ( -25.40, z-score = -2.64, R) >droYak2.chr2L 16287957 111 + 22324452 UAAAUAAUUGGAUUCUCAACGCAAUCGUCAUAGCCA-AAGUCAAUUAGCAGUCCUACAACUGGCUAAUC----GUAGCCGGAAAAAGCCCAGCGGAGAUUAGCCAAGUGUCCAAUA ......(((((((.((....(((((.(.(.......-..).).))).))...........(((((((((----...((.((.......)).))...))))))))))).))))))). ( -27.50, z-score = -2.54, R) >droSec1.super_3 2407785 79 - 7220098 ---------------------------------CUAUGAUUCAAUUAACAGUCCUACAACCGGCUAAUC----GCAGCCGGAAAAAGCCCAGCGGAGAUUAGCCAAGUGUCCAAUA ---------------------------------......................(((...((((((((----...((.((.......)).))...))))))))...)))...... ( -19.10, z-score = -2.04, R) >droSim1.chr2L 6668570 91 - 22036055 --------------------CCAUGAAUCCCAGCUA-AAAGCAAUUAACAGUCCUACAACUGGCUAAUC----GCAGCCGGAAAAAGCCCAGCGGAGAUUAGCCAAGUGUCCAAUA --------------------............((..-...)).............(((..(((((((((----...((.((.......)).))...)))))))))..)))...... ( -22.80, z-score = -2.41, R) >consensus ____________________CCA_CCUUCUUAGCUA_AAAUCAAUUAACAGUCCUACAACUGGCUAAUC____GCAGCCGGAAAAAGCCCAGCGGAGAUUAGCCAAGUGUCCAAUA .............................................................((((((((.......((.((.......)).))...))))))))............ ( -6.58 = -8.03 + 1.45)


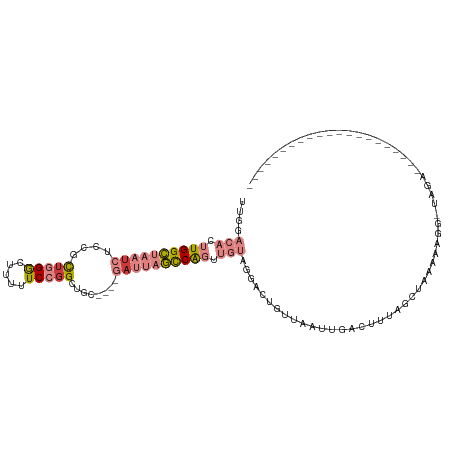
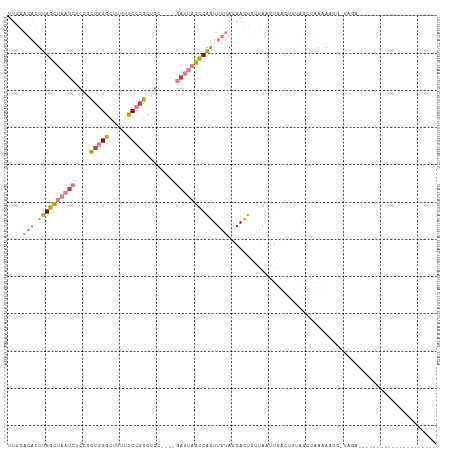
| Location | 6,867,673 – 6,867,764 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 59.69 |
| Shannon entropy | 0.69684 |
| G+C content | 0.42340 |
| Mean single sequence MFE | -24.44 |
| Consensus MFE | -9.12 |
| Energy contribution | -10.76 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.819401 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 6867673 91 + 23011544 UUGGACACUUGGCUAAUCUCCACUGGGCUUUUUCCGGCUGC----GAUUAGCCGGUUGUAGGACUGUUAAUUGACUUUAGCUGGGAAACAUGGAA-------------------- ....(((.((((((((((.(..(((((.....)))))..).----)))))))))).)))....(.(((((......))))).)(....)......-------------------- ( -25.20, z-score = -0.97, R) >droPer1.super_5 4862913 93 + 6813705 -----UUUUUGGGCAAU-UCCAUUAGACAAUAUCCGGCAGAAAGGGAAAUUUCAUUUCUAAGAAUGUCAGUUGAUUAUAGAAAAAAAGG-UAGUGUUGAU--------------- -----....((((....-)))).....((((((((....((((......)))).((((((.....(((....)))..))))))....))-..))))))..--------------- ( -13.10, z-score = 0.72, R) >dp4.chr4_group5 365402 94 + 2436548 -----UUUUUGGGCAAA-UCCAUUAGACAAUGUCCGGCAGAAAGGGAAAAUUCAUUUCUAAGAAUGCCAGUUGAUUAAAGAAAAAAAGGGUAGUGUUGAU--------------- -----....((((....-))))...((((.(..((((((.....(((((.....))))).....))))..((.....))........))..).))))...--------------- ( -16.20, z-score = 0.06, R) >droEre2.scaffold_4929 15786543 85 + 26641161 UUGGACACUUGGCUAAUCUCCGCUGGGCUCUUUCCUGCUAC----GAUUAGCCAGUUGUAGGACU--UAGCUAAC---AGCUAAAAUUGUUAAA--------------------- ....(((.((((((((((...((.(((.....))).))...----)))))))))).))).....(--((((....---.)))))..........--------------------- ( -25.00, z-score = -1.93, R) >droYak2.chr2L 16287959 111 - 22324452 UUGGACACUUGGCUAAUCUCCGCUGGGCUUUUUCCGGCUAC----GAUUAGCCAGUUGUAGGACUGCUAAUUGACUUUGGCUAUGACGAUUGCGUUGAGAAUCCAAUUAUUUAAC (((((..(((((((((((...((((((.....))))))...----))))))))((((((......(((((......)))))....)))))).....)))..)))))......... ( -34.70, z-score = -3.37, R) >droSec1.super_3 2407787 79 + 7220098 UUGGACACUUGGCUAAUCUCCGCUGGGCUUUUUCCGGCUGC----GAUUAGCCGGUUGUAGGACUGUUAAUUGAAUC---AUAGAA----------------------------- ....(((.((((((((((.(.((((((.....)))))).).----)))))))))).)))..................---......----------------------------- ( -25.50, z-score = -2.15, R) >droSim1.chr2L 6668572 91 + 22036055 UUGGACACUUGGCUAAUCUCCGCUGGGCUUUUUCCGGCUGC----GAUUAGCCAGUUGUAGGACUGUUAAUUGCUUUUAGCUGGGAUUCAUGGAA-------------------- .((((..((..(((((.....((((((.....)))))).((----(((((((.((((....)))))))))))))..)))))..)).)))).....-------------------- ( -31.40, z-score = -2.37, R) >consensus UUGGACACUUGGCUAAUCUCCGCUGGGCUUUUUCCGGCUGC____GAUUAGCCAGUUGUAGGACUGUUAAUUGACUUUAGCUAAAAAGG_UAGA_____________________ ....(((.((((((((((....(((((.....)))))........)))))))))).)))........................................................ ( -9.12 = -10.76 + 1.64)

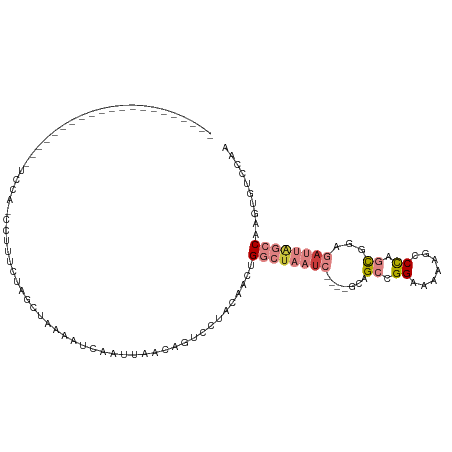
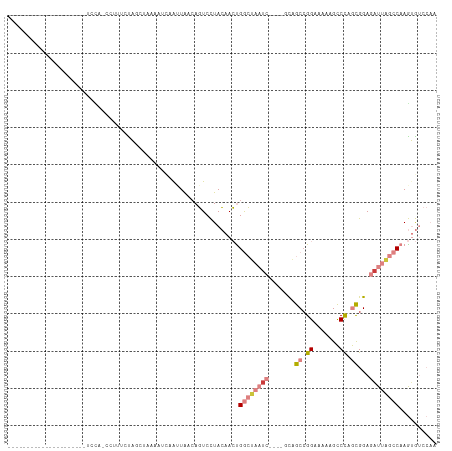
| Location | 6,867,673 – 6,867,764 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 59.69 |
| Shannon entropy | 0.69684 |
| G+C content | 0.42340 |
| Mean single sequence MFE | -19.16 |
| Consensus MFE | -6.58 |
| Energy contribution | -8.03 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.738115 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 6867673 91 - 23011544 --------------------UUCCAUGUUUCCCAGCUAAAGUCAAUUAACAGUCCUACAACCGGCUAAUC----GCAGCCGGAAAAAGCCCAGUGGAGAUUAGCCAAGUGUCCAA --------------------.....((((.....((....)).....)))).....(((...((((((((----...((.((.......)).))...))))))))...))).... ( -18.70, z-score = -1.31, R) >droPer1.super_5 4862913 93 - 6813705 ---------------AUCAACACUA-CCUUUUUUUCUAUAAUCAACUGACAUUCUUAGAAAUGAAAUUUCCCUUUCUGCCGGAUAUUGUCUAAUGGA-AUUGCCCAAAAA----- ---------------..........-.....................((((.(((((((((.(.......).))))))..)))...))))...(((.-.....)))....----- ( -9.80, z-score = -0.08, R) >dp4.chr4_group5 365402 94 - 2436548 ---------------AUCAACACUACCCUUUUUUUCUUUAAUCAACUGGCAUUCUUAGAAAUGAAUUUUCCCUUUCUGCCGGACAUUGUCUAAUGGA-UUUGCCCAAAAA----- ---------------..............................((((((......((((.....))))......))))))...........(((.-.....)))....----- ( -12.30, z-score = -0.38, R) >droEre2.scaffold_4929 15786543 85 - 26641161 ---------------------UUUAACAAUUUUAGCU---GUUAGCUA--AGUCCUACAACUGGCUAAUC----GUAGCAGGAAAGAGCCCAGCGGAGAUUAGCCAAGUGUCCAA ---------------------.........((((((.---....))))--))....(((..(((((((((----...((.((.......)).))...)))))))))..))).... ( -25.40, z-score = -2.50, R) >droYak2.chr2L 16287959 111 + 22324452 GUUAAAUAAUUGGAUUCUCAACGCAAUCGUCAUAGCCAAAGUCAAUUAGCAGUCCUACAACUGGCUAAUC----GUAGCCGGAAAAAGCCCAGCGGAGAUUAGCCAAGUGUCCAA (((((....((((.......(((....))).....))))......)))))......(((..(((((((((----...((.((.......)).))...)))))))))..))).... ( -26.00, z-score = -1.86, R) >droSec1.super_3 2407787 79 - 7220098 -----------------------------UUCUAU---GAUUCAAUUAACAGUCCUACAACCGGCUAAUC----GCAGCCGGAAAAAGCCCAGCGGAGAUUAGCCAAGUGUCCAA -----------------------------......---..................(((...((((((((----...((.((.......)).))...))))))))...))).... ( -19.10, z-score = -1.96, R) >droSim1.chr2L 6668572 91 - 22036055 --------------------UUCCAUGAAUCCCAGCUAAAAGCAAUUAACAGUCCUACAACUGGCUAAUC----GCAGCCGGAAAAAGCCCAGCGGAGAUUAGCCAAGUGUCCAA --------------------..............((.....)).............(((..(((((((((----...((.((.......)).))...)))))))))..))).... ( -22.80, z-score = -2.34, R) >consensus _____________________UCCA_CCUUUCUAGCUAAAAUCAAUUAACAGUCCUACAACUGGCUAAUC____GCAGCCGGAAAAAGCCCAGCGGAGAUUAGCCAAGUGUCCAA ..............................................................((((((((.......((.((.......)).))...)))))))).......... ( -6.58 = -8.03 + 1.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:22:25 2011