| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,233,836 – 6,233,967 |
| Length | 131 |
| Max. P | 0.998057 |

| Location | 6,233,836 – 6,233,932 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 76.96 |
| Shannon entropy | 0.49606 |
| G+C content | 0.34932 |
| Mean single sequence MFE | -21.53 |
| Consensus MFE | -18.56 |
| Energy contribution | -18.56 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.79 |
| SVM RNA-class probability | 0.995284 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
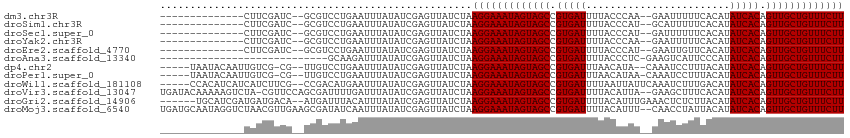
>dm3.chr3R 6233836 96 + 27905053 --------------CUUCGAUC--GCGUCCUGAAUUUAUAUCGAGUUAUCUAAGGAAAUAGUAGCCGUGAUUUUACCCAA--GAAUUUUUCACAUAUCACAGUUGCUGUUUCUU --------------.((((((.--...............)))))).......(((((((((((((.(((((.........--.............))))).))))))))))))) ( -20.74, z-score = -2.14, R) >droSim1.chr3R 12357971 96 - 27517382 --------------CUUCGAUC--GCGUCCUGAAUUUAUAUCGAGUUAUCUAAGGAAAUAGUAGCCGUGAUUUUACCCAU--GCAUUUUUCACAUAUCACAGUUGCUGUUUCUU --------------.((((((.--...............)))))).......(((((((((((((.(((((.........--.............))))).))))))))))))) ( -20.74, z-score = -2.12, R) >droSec1.super_0 5411074 96 - 21120651 --------------CUUCGAUC--GCGUCCUGAAUUUAUAUCGAGUUAUCUAAGGAAAUAGUAGCCGUGAUUUUACCCAU--GAUUUUUUCACAUAUCACAGUUGCUGUUUCUU --------------.((((((.--...............)))))).......(((((((((((((.(((((........(--((.....)))...))))).))))))))))))) ( -21.49, z-score = -2.39, R) >droYak2.chr3R 10262351 96 + 28832112 --------------CUUCGAUC--GCGUCCUGAAUUUAUAUCGAGUUAUCUAAGGAAAUAGUAGCCGUGAUUUUACCCAA--GAAUUUUUCACAUAUCACAGUUGCUGUUUCUU --------------.((((((.--...............)))))).......(((((((((((((.(((((.........--.............))))).))))))))))))) ( -20.74, z-score = -2.14, R) >droEre2.scaffold_4770 2362406 96 - 17746568 --------------CUUCGAUC--GCGUCCUGAAUUUAUAUCGAGUUAUCUAAGGAAAUAGUAGCCGUGAUUUUACCCAU--GAAUUGUUCACAUAUCACAGUUGCUGUUUCUU --------------.((((((.--...............)))))).......(((((((((((((.(((((......((.--....)).......))))).))))))))))))) ( -21.61, z-score = -2.01, R) >droAna3.scaffold_13340 22830288 85 - 23697760 ----------------------------GCAAGAUUUAUAUCGAGUUAUCUAAGGAAAUAGUAGCCGUGAUUUUACCCUC-GAAGUCAUUCCCAUAUCACAGUUGCUGUUUCUU ----------------------------....(((....)))..........(((((((((((((.(((((.........-(((....)))....))))).))))))))))))) ( -19.32, z-score = -2.84, R) >dp4.chr2 23560567 104 - 30794189 -----UAAUACAAUUGUCG-CG--UUGUCCUGAAUUUAUAUCGAGUUAUCUAAGGAAAUAGUAGCCGUGAUUUAACAUA--CAAAUCCUUUACAUAUCACAGUUGCUGUUUCUU -----....(((((.....-.)--))))........................(((((((((((((.(((((........--..............))))).))))))))))))) ( -19.75, z-score = -1.48, R) >droPer1.super_0 10728677 105 - 11822988 -----UAAUACAAUUGUCG-CG--UUGUCCUGAAUUUAUAUCGAGUUAUCUAAGGAAAUAGUAGCCGUGAUUUAACAUAA-CAAAUCCUUUACAUAUCACAGUUGCUGUUUCUU -----....(((((.....-.)--))))........................(((((((((((((.(((((.........-..............))))).))))))))))))) ( -19.70, z-score = -1.48, R) >droWil1.scaffold_181108 477 107 + 4707319 -----CCACAUCAUCAUCUUCG--CCGACAUGAAUUUAUAUCGAGUUAUCUAAGGAAAUAGUAGCCGUGAUUUUAAUUAUUCAAAUCUUUGACAUAUCACAGUUGCUGUUUCUU -----................(--((((.(((....))).))).))......(((((((((((((.(((((.((((............))))...))))).))))))))))))) ( -21.20, z-score = -2.07, R) >droVir3.scaffold_13047 1194139 111 + 19223366 UGAUACAAAAAGUCUA-CGUUCCAGCGAUUUUGAUUUAUAUCGAGUUAUCUAAGGAAAUAGUAGCCGUGAUUUUACAUUA--GAAGCUUUCACAUAUCACAGUUGCUGUUUCUU .((((...........-(((....)))..((((((....)))))).))))..(((((((((((((.(((((.........--.............))))).))))))))))))) ( -24.05, z-score = -1.82, R) >droGri2.scaffold_14906 9095247 106 - 14172833 ------UGCAUCGAUGAUGACA--AUGAUUUACAUUUAUAUCGAGUUAUCUAAGGAAAUAGUAGCCGUGAUUUUACAUUUGAAACUCUCUUACAUAUCACAGUUGCUGUUUCUU ------.........((((((.--.((((.((....)).)))).))))))..(((((((((((((.(((((.........(.....)........))))).))))))))))))) ( -24.13, z-score = -2.03, R) >droMoj3.scaffold_6540 5225146 112 + 34148556 UGAUGCAAUAGGUCUAACGUUGAAGCGAUAUCAAUUUAUAUCGAGUUAUCUAAGGAAAUAGUAGCCGUGAUUUUACAUUU--CAACCUAUUACAUAUCACAGUUGCUGUUUCUU ..................(.(((..((((((......))))))..))).)..(((((((((((((.(((((.........--.............))))).))))))))))))) ( -24.85, z-score = -1.87, R) >consensus ______________CUUCGAUC__GCGUCCUGAAUUUAUAUCGAGUUAUCUAAGGAAAUAGUAGCCGUGAUUUUACCCAU__GAAUCUUUCACAUAUCACAGUUGCUGUUUCUU ....................................................(((((((((((((.(((((........................))))).))))))))))))) (-18.56 = -18.56 + 0.00)
| Location | 6,233,858 – 6,233,967 |
|---|---|
| Length | 109 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 87.01 |
| Shannon entropy | 0.29309 |
| G+C content | 0.35363 |
| Mean single sequence MFE | -33.72 |
| Consensus MFE | -33.16 |
| Energy contribution | -33.16 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -5.38 |
| Structure conservation index | 0.98 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.25 |
| SVM RNA-class probability | 0.998057 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
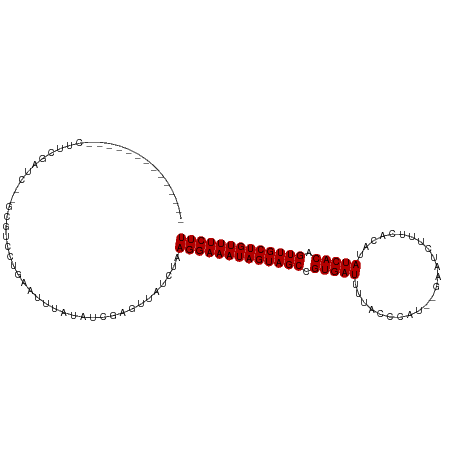
>dm3.chr3R 6233858 109 + 27905053 UAUCGAGUUAUCUAAGGAAAUAGUAGCCGUGAUUUUACCCAA-GAA-UUUUUCACAUAUCACAGUUGCUGUUUCUUUUAGAUAGCUCUUUUGUAAACAGCCUAGAUCCGAA---- ....(((((((((((((((((((((((.(((((.........-...-..........))))).))))))))))).))))))))))))........................---- ( -33.25, z-score = -5.54, R) >droSim1.chr3R 12357993 109 - 27517382 UAUCGAGUUAUCUAAGGAAAUAGUAGCCGUGAUUUUACCCAU-GCA-UUUUUCACAUAUCACAGUUGCUGUUUCUUUUAGAUAGCUCUUUUGUAAGCAGCCUAGAUCCGAA---- .((((((((((((((((((((((((((.(((((.........-...-..........))))).))))))))))).))))))))))))..(((....)))....))).....---- ( -33.35, z-score = -5.14, R) >droSec1.super_0 5411096 109 - 21120651 UAUCGAGUUAUCUAAGGAAAUAGUAGCCGUGAUUUUACCCAU-GAU-UUUUUCACAUAUCACAGUUGCUGUUUCUUUUAGAUAGCUCUUUUGUAAGCAGCCUAGAUCCGAA---- .((((((((((((((((((((((((((.(((((........(-((.-....)))...))))).))))))))))).))))))))))))..(((....)))....))).....---- ( -34.10, z-score = -5.49, R) >droYak2.chr3R 10262373 109 + 28832112 UAUCGAGUUAUCUAAGGAAAUAGUAGCCGUGAUUUUACCCAA-GAA-UUUUUCACAUAUCACAGUUGCUGUUUCUUUUAGAUAGCUCUUUUGUAAGCAGCCUAGUUCCGAA---- ....(((((((((((((((((((((((.(((((.........-...-..........))))).))))))))))).))))))))))))........................---- ( -33.25, z-score = -5.35, R) >droEre2.scaffold_4770 2362428 109 - 17746568 UAUCGAGUUAUCUAAGGAAAUAGUAGCCGUGAUUUUACCCAU-GAA-UUGUUCACAUAUCACAGUUGCUGUUUCUUUUAGAUAGCUCUUUUGUAAGCAGCCUAGUUCCGAA---- ....(((((((((((((((((((((((.(((((......((.-...-.)).......))))).))))))))))).))))))))))))........................---- ( -34.12, z-score = -5.25, R) >droAna3.scaffold_13340 22830298 110 - 23697760 UAUCGAGUUAUCUAAGGAAAUAGUAGCCGUGAUUUUACCCUC-GAAGUCAUUCCCAUAUCACAGUUGCUGUUUCUUUUAGAUAGCUCUUUUGUAAGCAGCCUAGAUUUCCA---- .((((((((((((((((((((((((((.(((((.........-(((....)))....))))).))))))))))).))))))))))))..(((....)))....))).....---- ( -33.42, z-score = -5.55, R) >dp4.chr2 23560597 108 - 30794189 UAUCGAGUUAUCUAAGGAAAUAGUAGCCGUGAUUUAACAUA--CAAAUCCUUUACAUAUCACAGUUGCUGUUUCUUUUAGAUAGCUCUUUUGUAAGCAGCCUAGAUCGGG----- ....(((((((((((((((((((((((.(((((........--..............))))).))))))))))).))))))))))))............((......)).----- ( -34.05, z-score = -4.84, R) >droPer1.super_0 10728707 109 - 11822988 UAUCGAGUUAUCUAAGGAAAUAGUAGCCGUGAUUUAACAUAA-CAAAUCCUUUACAUAUCACAGUUGCUGUUUCUUUUAGAUAGCUCUUUUGUAAGCAGCCUAGAUCGGG----- ....(((((((((((((((((((((((.(((((.........-..............))))).))))))))))).))))))))))))............((......)).----- ( -34.00, z-score = -4.83, R) >droWil1.scaffold_181108 508 103 + 4707319 UAUCGAGUUAUCUAAGGAAAUAGUAGCCGUGAUUUUAAUUAUUCAAAUCUUUGACAUAUCACAGUUGCUGUUUCUUUUAGAUAGCUCUUUUGUAAGCAGCCUA------------ ....(((((((((((((((((((((((.(((((.((((............))))...))))).))))))))))).))))))))))))................------------ ( -34.00, z-score = -5.79, R) >droVir3.scaffold_13047 1194176 102 + 19223366 UAUCGAGUUAUCUAAGGAAAUAGUAGCCGUGAUUUUACAUUA--GAAGCUUUCACAUAUCACAGUUGCUGUUUCUUUUAGAUAGCUCUUUUGUAAGCAGCCUAA----------- ....(((((((((((((((((((((((.(((((.........--.............))))).))))))))))).)))))))))))).................----------- ( -33.25, z-score = -4.99, R) >droGri2.scaffold_14906 9095277 115 - 14172833 UAUCGAGUUAUCUAAGGAAAUAGUAGCCGUGAUUUUACAUUUGAAACUCUCUUACAUAUCACAGUUGCUGUUUCUUUUAGAUAGCUCUUUUGUAAGCAACAAACCCUAACUCCUC ....(((((((((((((((((((((((.(((((.........(.....)........))))).))))))))))).)))))))))))).(((((.....)))))............ ( -34.63, z-score = -5.80, R) >droMoj3.scaffold_6540 5225184 102 + 34148556 UAUCGAGUUAUCUAAGGAAAUAGUAGCCGUGAUUUUACAUUUCAA--CCUAUUACAUAUCACAGUUGCUGUUUCUUUUAGAUAGCUCUUUUGUAAGCAGCCUAA----------- ....(((((((((((((((((((((((.(((((............--..........))))).))))))))))).)))))))))))).................----------- ( -33.25, z-score = -5.96, R) >consensus UAUCGAGUUAUCUAAGGAAAUAGUAGCCGUGAUUUUACCCAU_GAA_UCUUUCACAUAUCACAGUUGCUGUUUCUUUUAGAUAGCUCUUUUGUAAGCAGCCUAGAUCCGAA____ ....(((((((((((((((((((((((.(((((........................))))).))))))))))).))))))))))))............................ (-33.16 = -33.16 + 0.00)

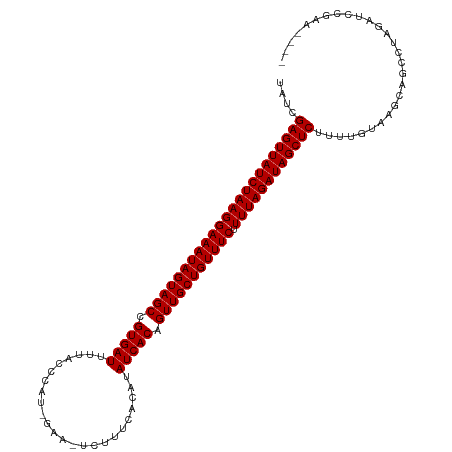
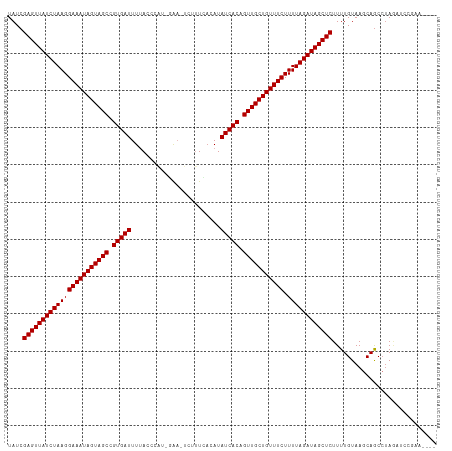
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:02:50 2011