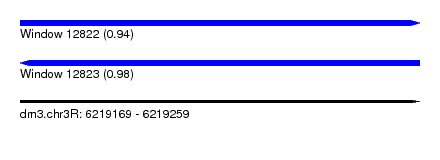
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,219,169 – 6,219,259 |
| Length | 90 |
| Max. P | 0.976984 |

| Location | 6,219,169 – 6,219,259 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 72.71 |
| Shannon entropy | 0.50966 |
| G+C content | 0.50256 |
| Mean single sequence MFE | -17.57 |
| Consensus MFE | -9.48 |
| Energy contribution | -10.37 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.941099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
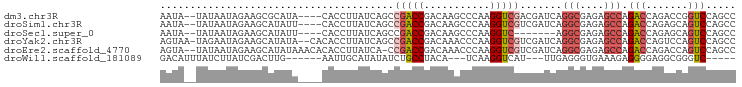
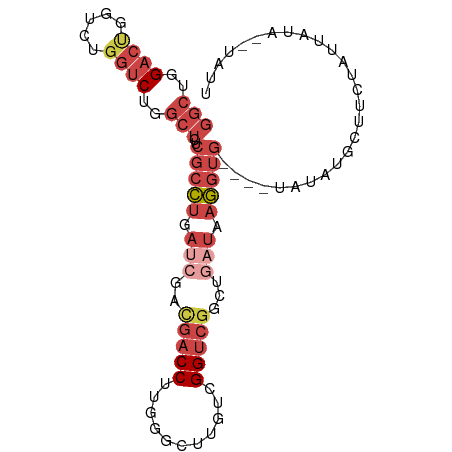
>dm3.chr3R 6219169 90 + 27905053 AAUA--UAUAAUAGAAGCGCAUA----CACCUUAUCAGCCGACCGACAAGCCCAAGGUCGACGAUCAGGCGAGAGCCAGACCAGACCGGUCCAGCC ....--............((...----..........(.(((((...........))))).).....(((....))).((((.....))))..)). ( -20.80, z-score = -2.23, R) >droSim1.chr3R 12339216 90 - 27517382 AAUA--UAUAAUAGAAGCAUAUU----CACCUUAUCAGCCGACCGACAAGCCCAAGGUCGUCGAUCAGGCGAGAGCCAGACCAGAGCAGUCCAGCC ....--...............((----(.(((.(((.(.(((((...........))))).)))).))).))).((..(((.......)))..)). ( -18.30, z-score = -2.00, R) >droSec1.super_0 5394417 83 - 21120651 AAUA--UAUAAUAGAAGCAUAUU----CACCUUAUCAGCCGACCGACAAGCCCAAGGUC-------AGGCGAGAGCCAGACCAGAGCAGUCCAGCC ....--..........((.....----...(((.((........)).))).....((((-------.(((....))).))))...))......... ( -16.90, z-score = -2.72, R) >droYak2.chr3R 10245001 93 + 28832112 AGUAA-UAGAAUAGAAGCAUAUA--CACACCUUAUCAGCCGACCGACAAACCCAAGGUCGUCGAUCAGGCGAGAGCCAGACCAGUCCAGUCCAGCC .....-..........((.....--............(.(((((...........))))).)(.((.(((....))).)).)...........)). ( -16.60, z-score = -1.62, R) >droEre2.scaffold_4770 2345349 93 - 17746568 AGUA--UAUAAUAGAAGCAUAUAAACACACCUUAUCA-CCGACCGACAAACCCAAGGUCGUCGAUCAGGCGAGAGCCAGACCAGACCAGUCCAGCC ....--..........((...............(((.-.(((((...........)))))..)))..(((....))).(((.......)))..)). ( -17.90, z-score = -2.83, R) >droWil1.scaffold_181089 12279869 79 - 12369635 GACAUUUAUCUUAUCGACUUG------AAUUGCAUAUAUCUGCCUACA---UCAAGGUCAU---UUGAGGGUGAAAGAGGGGAGGCGGGUC----- (((.....(((..((..((((------(...(((......))).....---))))).((((---(....)))))..))..))).....)))----- ( -14.90, z-score = 0.71, R) >consensus AAUA__UAUAAUAGAAGCAUAUA____CACCUUAUCAGCCGACCGACAAGCCCAAGGUCGUCGAUCAGGCGAGAGCCAGACCAGACCAGUCCAGCC .......................................(((((...........))))).......(((....))).(((.......)))..... ( -9.48 = -10.37 + 0.89)
| Location | 6,219,169 – 6,219,259 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 72.71 |
| Shannon entropy | 0.50966 |
| G+C content | 0.50256 |
| Mean single sequence MFE | -27.67 |
| Consensus MFE | -15.28 |
| Energy contribution | -17.78 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.976984 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
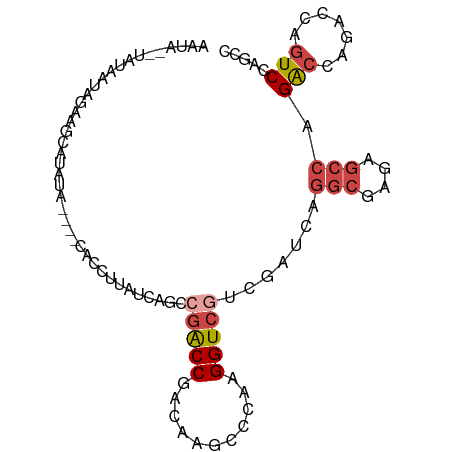
>dm3.chr3R 6219169 90 - 27905053 GGCUGGACCGGUCUGGUCUGGCUCUCGCCUGAUCGUCGACCUUGGGCUUGUCGGUCGGCUGAUAAGGUG----UAUGCGCUUCUAUUAUA--UAUU (((..((((.....))))..)))..(((((.((((((((((...........))))))).))).)))))----.................--.... ( -32.20, z-score = -1.79, R) >droSim1.chr3R 12339216 90 + 27517382 GGCUGGACUGCUCUGGUCUGGCUCUCGCCUGAUCGACGACCUUGGGCUUGUCGGUCGGCUGAUAAGGUG----AAUAUGCUUCUAUUAUA--UAUU (((..((((.....))))..)))...(((.(((((((((((...)).))))))))))))(((((((((.----.....)))).)))))..--.... ( -31.40, z-score = -2.73, R) >droSec1.super_0 5394417 83 + 21120651 GGCUGGACUGCUCUGGUCUGGCUCUCGCCU-------GACCUUGGGCUUGUCGGUCGGCUGAUAAGGUG----AAUAUGCUUCUAUUAUA--UAUU ((((((...((((.((((.(((....))).-------))))..))))...))))))...(((((((((.----.....)))).)))))..--.... ( -28.10, z-score = -2.45, R) >droYak2.chr3R 10245001 93 - 28832112 GGCUGGACUGGACUGGUCUGGCUCUCGCCUGAUCGACGACCUUGGGUUUGUCGGUCGGCUGAUAAGGUGUG--UAUAUGCUUCUAUUCUA-UUACU (((..((((.....))))..)))...(((.(((((((((((...)).))))))))))))(((((..(((.(--(....))...)))..))-))).. ( -31.00, z-score = -2.10, R) >droEre2.scaffold_4770 2345349 93 + 17746568 GGCUGGACUGGUCUGGUCUGGCUCUCGCCUGAUCGACGACCUUGGGUUUGUCGGUCGG-UGAUAAGGUGUGUUUAUAUGCUUCUAUUAUA--UACU (((..((((.....))))..))).(((((.(((((((((((...)).)))))))))))-)))...(((((((..(((......))).)))--)))) ( -34.00, z-score = -3.43, R) >droWil1.scaffold_181089 12279869 79 + 12369635 -----GACCCGCCUCCCCUCUUUCACCCUCAA---AUGACCUUGA---UGUAGGCAGAUAUAUGCAAUU------CAAGUCGAUAAGAUAAAUGUC -----...........................---.((((.((((---.....(((......)))...)------))))))).............. ( -9.30, z-score = 0.43, R) >consensus GGCUGGACUGGUCUGGUCUGGCUCUCGCCUGAUCGACGACCUUGGGCUUGUCGGUCGGCUGAUAAGGUG____UAUAUGCUUCUAUUAUA__UAUU (((..((((.....))))..)))..(((((.(((..(((((...........)))))...))).)))))........................... (-15.28 = -17.78 + 2.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:02:45 2011