
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 6,862,378 – 6,862,562 |
| Length | 184 |
| Max. P | 0.792130 |

| Location | 6,862,378 – 6,862,473 |
|---|---|
| Length | 95 |
| Sequences | 14 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.70 |
| Shannon entropy | 0.50475 |
| G+C content | 0.53643 |
| Mean single sequence MFE | -30.89 |
| Consensus MFE | -18.81 |
| Energy contribution | -19.06 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.681002 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
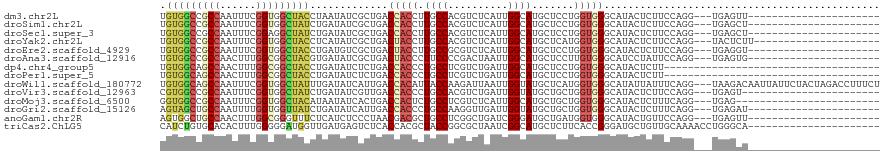
>dm3.chr2L 6862378 95 - 23011544 UGUGGCCGCCAAUUUCGGUGGCUACCUAAUAUCGCUGACCACCUUGCCACGUCUCAUUGGCAUGCUCCUGGUGGGCAUACUCUUCCAGG---UGAGUU---------------------- .(((((((((......)))))))))........(((.((((...(((((........)))))......)))).)))..((((.......---.)))).---------------------- ( -35.40, z-score = -2.27, R) >droSim1.chr2L 6662747 95 - 22036055 UGUGGCCGCCAAUUUCGGUGGCUAUCUGAUAUCGCUGACCACCUUGCCACGUCUCAUUGGCAUGCUCCUGGUGGGCAUACUCUUCCAGG---UGAGCU---------------------- .(((((((((......)))))))))........(((.(((.....((((........))))((((.((....)))))).........))---).))).---------------------- ( -35.90, z-score = -1.98, R) >droSec1.super_3 2402527 95 - 7220098 UGUGGCCGCCAAUUUCGGAGGCUAUCUGAUAUCGCUGACCACCUUGCCACGUCUCAUUGGCAUGCUCCUGGUGGGCAUACUCUUCCAGG---UGAGCU---------------------- .((((((.((......)).))))))........(((.(((.....((((........))))((((.((....)))))).........))---).))).---------------------- ( -32.10, z-score = -0.72, R) >droYak2.chr2L 16282401 96 + 22324452 UGUGGCCGCCAAUUUCGGUGGCUACCUCAUAUCGCUGACUACCUUGCCACGUCUCAUUGGCAUGCUCAUGGUGGGCAUACUCUUCCAGG---UACUCUU--------------------- .(((((((((......)))))))))...........((.(((((.((((........))))((((((.....))))))........)))---)).))..--------------------- ( -34.20, z-score = -3.00, R) >droEre2.scaffold_4929 15781353 95 - 26641161 UGUGGCCGCCAAUUUCGGUGGCUACCUGAUGUCGCUGACUACCUUGCCGCGUCUCAUUGGCAUGCUCCUGGUGGGCAUACUCUUCCAGG---UGAGGU---------------------- .(((((((((......))))))))).....(((...))).((((..((..(((.....)))((((.((....)))))).........))---..))))---------------------- ( -36.70, z-score = -1.46, R) >droAna3.scaffold_12916 8449722 95 - 16180835 UGUGGCCGCCAACUUUGGCGGCUACGUGAUAUCGCUGACUACCCUUCCCCGACUAAUUGGCAUGCUCCUUGUGGGCAUCCUAUUCCAGG---UGAGUG---------------------- .((((((((((....))))))))))(((....))).........(((.((....(((.((.((((.((....)))))))).)))...))---.)))..---------------------- ( -33.80, z-score = -2.31, R) >dp4.chr4_group5 359883 84 - 2436548 UGUGGCAGCCAACUUUGGCGGCUACCUGAUAUCUCUGACCACCCUGCCUCGUCUGAUUGGCAUGCUCCUGGUGGGCAUACUCUU------------------------------------ .(((((.((((....)))).)))))..........((.(((((..((...(((.....)))..))....))))).)).......------------------------------------ ( -24.60, z-score = -0.35, R) >droPer1.super_5 4857432 84 - 6813705 UGUGGCAGCCAACUUUGGCGGCUACCUGAUAUCUCUGACCACCCUGCCUCGUCUGAUUGGCAUGCUCCUGGUGGGCAUACUCUU------------------------------------ .(((((.((((....)))).)))))..........((.(((((..((...(((.....)))..))....))))).)).......------------------------------------ ( -24.60, z-score = -0.35, R) >droWil1.scaffold_180772 2036934 117 + 8906247 UGUGGCAGCCAAUUUCGGUGGCUAUUUGAUAUCAUUGACCACAUUACCAAGAUUAAUUGGUAUGCUCAUGGUGGGCAUAUUAUUUCAGG---UAAGACAAUUAUUCUACUAGACCUUUCU (((((((((((.......)))))...((....))..).)))))(((((..((.....((((((((((.....))))))))))..)).))---)))......................... ( -26.40, z-score = -0.43, R) >droVir3.scaffold_12963 5400754 94 + 20206255 CGUGGCCGCCAAUUUCGGUGGCUAUCUGAUAUCGUUGACCACCCUGCCACGUCUGAUUGGUAUGCUGCUGGUGGGCAUACUCUUCCAGG---UGAGU----------------------- .(((((((((......)))))))))..........((.(((((..((..(((((....)).)))..)).))))).)).((((.......---.))))----------------------- ( -29.60, z-score = -0.01, R) >droMoj3.scaffold_6500 14595795 93 + 32352404 GGUGGCCGCCAAUUUCGGUGGCUACAUAAUAUCACUGACCACUCUGCCUCGUCUCAUUGGCAUGCUGCUGGUGGGCAUACUCUUUCAGG---UGAG------------------------ .(((((((((......)))))))))......(((((((......((((......(((..((.....))..))))))).......))).)---))).------------------------ ( -33.42, z-score = -2.22, R) >droGri2.scaffold_15126 4560131 95 - 8399593 AGUAGCUGCCAAUUUUGGUGGUUAUCUGAUAUCAUUGACCACCCUGCCAAGGUUGAUUGGUAUGCUGCUGGUGGGCAUACUCUUUCAGG---UGAGAU---------------------- ((((((((((((((..((((((((..((....)).))))))))..((....)).)))))))).))))))..........(((.......---.)))..---------------------- ( -30.70, z-score = -1.12, R) >anoGam1.chr2R 15237960 95 - 62725911 AGUGGCUGCCAACUUUGGCGGGUUUCUCAUCUCCCUAACGACGCUGCCUCGGCUGAUCGGGAUGCUGAUGGUGGGCAUACUGUUCCAGG---UGAGUU---------------------- .(..((.((((....))))(((..........))).......))..)((((.(((..(((.(((((.......))))).)))...))).---))))..---------------------- ( -29.90, z-score = 0.60, R) >triCas2.ChLG5 17199697 98 - 18847211 CAUCUGUGCACACUUUGGGGGAUGGUUGAUGAGUCUCACCACGCUACCGGCGCUAAUCGGCAUGCUCUUCACCGGGAUGCUGUUGCAAAACCUGGGCA---------------------- ......(((........(((((((((.((......)))))).((..((((......))))...))))))).(((((.(((....)))...))))))))---------------------- ( -25.20, z-score = 1.63, R) >consensus UGUGGCCGCCAAUUUCGGUGGCUACCUGAUAUCGCUGACCACCCUGCCACGUCUCAUUGGCAUGCUCCUGGUGGGCAUACUCUUCCAGG___UGAGCU______________________ .(((((((((......))))))))).............(((((.((((..........)))).......))))).............................................. (-18.81 = -19.06 + 0.24)

| Location | 6,862,393 – 6,862,513 |
|---|---|
| Length | 120 |
| Sequences | 14 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.03 |
| Shannon entropy | 0.47374 |
| G+C content | 0.57619 |
| Mean single sequence MFE | -45.01 |
| Consensus MFE | -23.03 |
| Energy contribution | -23.09 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.583680 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
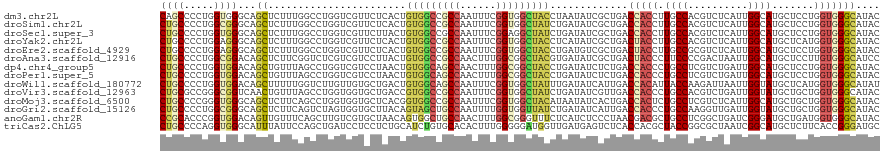
>dm3.chr2L 6862393 120 - 23011544 CAGCCCCUGGUGGGCAGCUCUUUGGCCUGGUCGUUCUCACUGUGGCCGCCAAUUUCGGUGGCUACCUAAUAUCGCUGACCACCUUGCCACGUCUCAUUGGCAUGCUCCUGGUGGGCAUAC ..(((((..(.(((((......((((.(((((((.......(((((((((......)))))))))........)).)))))....)))).(((.....))).))))))..).)))).... ( -49.86, z-score = -2.67, R) >droSim1.chr2L 6662762 120 - 22036055 CUGCCCCUGGCGGGCAGCUCUUUGGCCUGGUCGUUCUCACUGUGGCCGCCAAUUUCGGUGGCUAUCUGAUAUCGCUGACCACCUUGCCACGUCUCAUUGGCAUGCUCCUGGUGGGCAUAC .((((((..(.(((((......((((.(((((((..(((..(((((((((......))))))))).)))....)).)))))....)))).(((.....))).))))))..).)))))... ( -50.00, z-score = -2.75, R) >droSec1.super_3 2402542 120 - 7220098 CUGCCCCUGGUGGGCAGCUCUUUGGCCUGGUCGUUCUUACUGUGGCCGCCAAUUUCGGAGGCUAUCUGAUAUCGCUGACCACCUUGCCACGUCUCAUUGGCAUGCUCCUGGUGGGCAUAC .((((((((((((.((((...(((((..(((((.........))))))))))..(((((.....)))))....)))).))))).(((((........))))).......)).)))))... ( -45.70, z-score = -1.56, R) >droYak2.chr2L 16282417 120 + 22324452 CUGCCCCUGGAGGGCAGCUCUUUGGCCUGGUCGUUCUCACUGUGGCCGCCAAUUUCGGUGGCUACCUCAUAUCGCUGACUACCUUGCCACGUCUCAUUGGCAUGCUCAUGGUGGGCAUAC .((((((((..(((((......((((..(((.(((......(((((((((......)))))))))...........))).)))..)))).(((.....))).))))).))).)))))... ( -44.73, z-score = -1.46, R) >droEre2.scaffold_4929 15781368 120 - 26641161 CUGCCCCUGGAGGGCAGCUCUUUGGCCUGGUCGUUCUCACUGUGGCCGCCAAUUUCGGUGGCUACCUGAUGUCGCUGACUACCUUGCCGCGUCUCAUUGGCAUGCUCCUGGUGGGCAUAC .((((((..(.(((((.......(((..(((.(((..(((.(((((((((......))))))))).....)).)..))).)))..)))..(((.....))).))))))..).)))))... ( -49.70, z-score = -2.16, R) >droAna3.scaffold_12916 8449737 120 - 16180835 CUGCCCCUGGCGGACAGCUCUUCGGUCUCGUCGUCCUUACUGUGGCCGCCAACUUUGGCGGCUACGUGAUAUCGCUGACUACCCUUCCCCGACUAAUUGGCAUGCUCCUUGUGGGCAUCC .((((((.((.(..(((((..(..((((((.((...((((.((((((((((....))))))))))))))...)).)))............)))..)..))).))..))).).)))))... ( -42.50, z-score = -2.32, R) >dp4.chr4_group5 359887 120 - 2436548 CUGCCCCUGGUGGACAGCUGUUUAGCCUGGUCGUCCUAACUGUGGCAGCCAACUUUGGCGGCUACCUGAUAUCUCUGACCACCCUGCCUCGUCUGAUUGGCAUGCUCCUGGUGGGCAUAC .((((((((((((.(((....((((...((....)).....(((((.((((....)))).))))))))).....))).))))).((((..........)))).......)).)))))... ( -39.90, z-score = -0.25, R) >droPer1.super_5 4857436 120 - 6813705 CUGCCCCUGGUGGACAGCUGUUUAGCCUGGUCGUCCUAACUGUGGCAGCCAACUUUGGCGGCUACCUGAUAUCUCUGACCACCCUGCCUCGUCUGAUUGGCAUGCUCCUGGUGGGCAUAC .((((((((((((.(((....((((...((....)).....(((((.((((....)))).))))))))).....))).))))).((((..........)))).......)).)))))... ( -39.90, z-score = -0.25, R) >droWil1.scaffold_180772 2036971 120 + 8906247 CUGCCCCUGGUGGACAGCUUUUUGGUCUUGUUGUGCUGACUGUGGCAGCCAAUUUCGGUGGCUAUUUGAUAUCAUUGACCACAUUACCAAGAUUAAUUGGUAUGCUCAUGGUGGGCAUAU .(((((.(.(((..(((((..((((((((((((.((((.(....)))))))).....(((((....((....))..).)))).....)))))))))..))).))..))).).)))))... ( -38.30, z-score = -1.09, R) >droVir3.scaffold_12963 5400768 120 + 20206255 CUGCGCCGGGCGGUCAACUGUUUAGCCUGGUGGUGCUGACCGUGGCCGCCAAUUUCGGUGGCUAUCUGAUAUCGUUGACCACCCUGCCACGUCUGAUUGGUAUGCUGCUGGUGGGCAUAC .((.((.(((.(((((((.(((..(((....)))...))).(((((((((......)))))))))........))))))).))).)))).((((.((..((.....))..)))))).... ( -48.30, z-score = -1.32, R) >droMoj3.scaffold_6500 14595808 120 + 32352404 CUGCCCCGGGUGGGCAGCUCUUCAGCCUGGUGGUGCUCACGGUGGCCGCCAAUUUCGGUGGCUACAUAAUAUCACUGACCACUCUGCCUCGUCUCAUUGGCAUGCUGCUGGUGGGCAUAC .(((((.....)))))((((.(((((.((((((((......(((((((((......))))))))).......)))).))))...((((..........))))....))))).)))).... ( -49.72, z-score = -1.68, R) >droGri2.scaffold_15126 4560146 120 - 8399593 CUGCCCCUGGCGGGCAGCUCUUCAGUCUAGUGGUGCUUACAGUAGCUGCCAAUUUUGGUGGUUAUCUGAUAUCAUUGACCACCCUGCCAAGGUUGAUUGGUAUGCUGCUGGUGGGCAUAC ((((((.....))))))...............((((((((((((((((((((((..((((((((..((....)).))))))))..((....)).)))))))).)))))).)))))))).. ( -49.50, z-score = -2.42, R) >anoGam1.chr2R 15237975 120 - 62725911 CCGCACCCGGUGGACAGUUGUUUCAGCUUGUCGUGCUAACAGUGGCUGCCAACUUUGGCGGGUUUCUCAUCUCCCUAACGACGCUGCCUCGGCUGAUCGGGAUGCUGAUGGUGGGCAUAC ..(((((((((...((((((...((((..(((((((((....)))).((((....))))(((..........)))..)))))))))...)))))))))))).)))............... ( -41.10, z-score = 0.07, R) >triCas2.ChLG5 17199715 120 - 18847211 CUGCCCCAGGUGGGCAUUUAUUCCAGCUGAUCCUCCUCUGCAUCUGUGCACACUUUGGGGGAUGGUUGAUGAGUCUCACCACGCUACCGGCGCUAAUCGGCAUGCUCUUCACCGGGAUGC ....(((.((((((.((((((..(((((.(((((((..((((....))))......))))))))))))))))))))))))..((..((((......))))...))........))).... ( -40.90, z-score = -0.31, R) >consensus CUGCCCCUGGUGGGCAGCUCUUUGGCCUGGUCGUGCUCACUGUGGCCGCCAAUUUCGGUGGCUACCUGAUAUCGCUGACCACCCUGCCACGUCUCAUUGGCAUGCUCCUGGUGGGCAUAC ((((.....))))...((.......................(((((((((......))))))))).............(((((.((((..........)))).......))))))).... (-23.03 = -23.09 + 0.06)
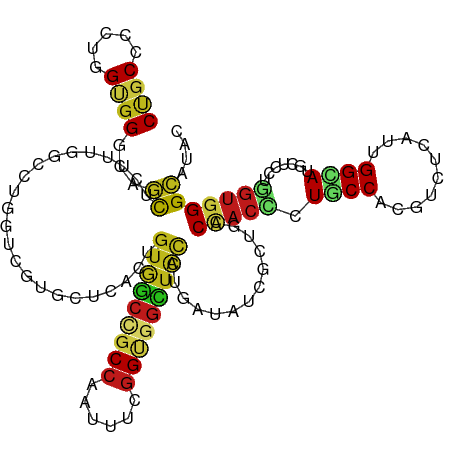

| Location | 6,862,443 – 6,862,562 |
|---|---|
| Length | 119 |
| Sequences | 14 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 77.17 |
| Shannon entropy | 0.53468 |
| G+C content | 0.55800 |
| Mean single sequence MFE | -38.82 |
| Consensus MFE | -14.60 |
| Energy contribution | -14.11 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.59 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.515598 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 6862443 119 + 23011544 AUUAGGUAGCCACCGAAAUUGGCGGCCACAGUGAGAACGACCAGGCCAAAGAGCUGCCCACCAGGGGCUGCCGAAUCGCCGAUGAUCACAAAGGCGGUGACCCAGAUAAGGACACCUAU ..(((((..((...((.((((((((((...((....)).....))))...(.((.((((.....)))).))).....))))))..)).....((.(....)))......))..))))). ( -41.40, z-score = -2.31, R) >droSim1.chr2L 6662812 119 + 22036055 AUCAGAUAGCCACCGAAAUUGGCGGCCACAGUGAGAACGACCAGGCCAAAGAGCUGCCCGCCAGGGGCAGCCGAGUCGCCGAUGAUCACAAAGGCGGUGACCCAGAUAAGGACACCUAU ........(((...((.((((((((((...((....)).....))))...(.(((((((.....)))))))).....))))))..)).....)))((((.((.......)).))))... ( -42.60, z-score = -2.82, R) >droSec1.super_3 2402592 119 + 7220098 AUCAGAUAGCCUCCGAAAUUGGCGGCCACAGUAAGAACGACCAGGCCAAAGAGCUGCCCACCAGGGGCAGCCGAGUCGCCGAUGAUCACAAAGGCGGUGACCCAGAUAAGGACUCCUAU .....((((..(((((.((((((((((...((....)).....))))...(.(((((((.....)))))))).....))))))..)).....((.(....)))......)))...)))) ( -40.40, z-score = -2.03, R) >droYak2.chr2L 16282467 119 - 22324452 AUGAGGUAGCCACCGAAAUUGGCGGCCACAGUGAGAACGACCAGGCCAAAGAGCUGCCCUCCAGGGGCAGCCGAGUCGCCGAUGAUCACGAAGGCGGUGACCCAGAUAAGGACACCUGU ...((((..((...((.((((((((((...((....)).....))))...(.(((((((.....)))))))).....))))))..)).....((.(....)))......))..)))).. ( -45.40, z-score = -2.37, R) >droEre2.scaffold_4929 15781418 119 + 26641161 AUCAGGUAGCCACCGAAAUUGGCGGCCACAGUGAGAACGACCAGGCCAAAGAGCUGCCCUCCAGGGGCAGCCGAGUCGCCGAUGAUCACAAAGGCGGUGACCCAGAUGAGGACACCUGU ..(((((..((...((.((((((((((...((....)).....))))...(.(((((((.....)))))))).....))))))..)).....((.(....)))......))..))))). ( -47.60, z-score = -2.75, R) >droAna3.scaffold_12916 8449787 119 + 16180835 AUCACGUAGCCGCCAAAGUUGGCGGCCACAGUAAGGACGACGAGACCGAAGAGCUGUCCGCCAGGGGCAGCCGAGUCGCCAAUGAUUACAAAGGCUGUUAUCCAAAUAAGGACACCUGU .....((.(((((((....))))))).)).....(((..(((.(((((....(((((((.....))))))))).)))(((..((....))..))))))..))).....(((...))).. ( -42.70, z-score = -2.67, R) >dp4.chr4_group5 359937 119 + 2436548 AUCAGGUAGCCGCCAAAGUUGGCUGCCACAGUUAGGACGACCAGGCUAAACAGCUGUCCACCAGGGGCAGCCGAGUCUCCAAUGAUUACAAAGGCCGUGAUCCAAAUGAGGACACCUGA .((((((...........(((((((((.(.....((((.....((((....)))))))).....))))))))))(((..((..((((((.......))))))....))..))))))))) ( -42.50, z-score = -2.09, R) >droPer1.super_5 4857486 119 + 6813705 AUCAGGUAGCCGCCAAAGUUGGCUGCCACAGUUAGGACGACCAGGCUAAACAGCUGUCCACCAGGGGCAGCCGAGUCUCCAAUGAUUACAAAGGCCGUGAUCCAAAUGAGGACACCUGA .((((((...........(((((((((.(.....((((.....((((....)))))))).....))))))))))(((..((..((((((.......))))))....))..))))))))) ( -42.50, z-score = -2.09, R) >droWil1.scaffold_180772 2037021 119 - 8906247 AUCAAAUAGCCACCGAAAUUGGCUGCCACAGUCAGCACAACAAGACCAAAAAGCUGUCCACCAGGGGCAGCUGAGUCUCCAAUGAUCACAAAGGCCGUGACCCAUAUUAGGACACCUAC ......((((((.......)))))).....(((.........((((.....((((((((.....))))))))..))))...(((.((((.......))))..))).....)))...... ( -29.70, z-score = -1.07, R) >droVir3.scaffold_12963 5400818 119 - 20206255 AUCAGAUAGCCACCGAAAUUGGCGGCCACGGUCAGCACCACCAGGCUAAACAGUUGACCGCCCGGCGCAGCUGUGUCCCCGAUAAUCACAAAUGCCACAAUCCAAAGUAGCAAACCUGC ........((((.......)))).(((.((((((((................))))))))...)))((((.((((...........))))..(((.((........)).)))...)))) ( -29.49, z-score = -0.16, R) >droMoj3.scaffold_6500 14595858 118 - 32352404 AUUAUGUAGCCACCGAAAUUGGCGGCCACCGUGAGCACCACCAGGCUGAAGAGCUGCCCACCCGGGGCAGCUGUGUCCCCAAUGAUCACAAAGGCUACAAUCCAGAGUAGUAGACCUA- ....(((((((...((.((((((((((...(((.....)))..))))).(.((((((((.....)))))))).)....)))))..)).....)))))))...................- ( -40.90, z-score = -2.11, R) >droGri2.scaffold_15126 4560196 118 + 8399593 AUCAGAUAACCACCAAAAUUGGCAGCUACUGUAAGCACCACUAGACUGAAGAGCUGCCCGCCAGGGGCAGCGGUGUCCCCAAUAAUCACAAAUGCCACAAUCCAAAGUAGCAAACCUA- ..........((((....((..((((((.((.......)).))).)))..))(((((((.....))))))))))).................(((.((........)).)))......- ( -29.40, z-score = -1.19, R) >anoGam1.chr2R 15238025 116 + 62725911 AUGAGAAACCCGCCAAAGUUGGCAGCCACUGUUAGCACGACAAGCUGAAACAACUGUCCACCGGGUGCGGCAGUGUCACCGAUGAUGACAAACGCCGUUAUCCAUAGUAGUACACC--- (((.((.(((((....(((((.((((...((((.....)))).))))...)))))......)))))(((((..(((((.......)))))...)))))..)))))...........--- ( -33.70, z-score = -1.24, R) >triCas2.ChLG5 17199765 119 + 18847211 AUCAACCAUCCCCCAAAGUGUGCACAGAUGCAGAGGAGGAUCAGCUGGAAUAAAUGCCCACCUGGGGCAGCGGUGGCCCCGACGAUGGCGUAGAGGAUGCACCAGGAUAGCGUCCCGAU (((..((((((((((..(((.(((...((.(((..(.....)..)))..))...))).))).)))))..((((.....))).))))))....(.((((((.........)))))))))) ( -35.20, z-score = 1.06, R) >consensus AUCAGGUAGCCACCGAAAUUGGCGGCCACAGUGAGAACGACCAGGCCAAAGAGCUGCCCACCAGGGGCAGCCGAGUCGCCGAUGAUCACAAAGGCCGUGAUCCAGAUUAGGACACCUAU .................(((((........((....)).....(((......(((((((.....)))))))...))).))))).................................... (-14.60 = -14.11 + -0.48)

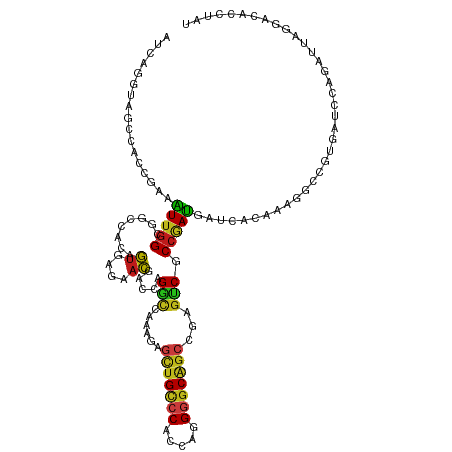
| Location | 6,862,443 – 6,862,562 |
|---|---|
| Length | 119 |
| Sequences | 14 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 77.17 |
| Shannon entropy | 0.53468 |
| G+C content | 0.55800 |
| Mean single sequence MFE | -45.88 |
| Consensus MFE | -20.06 |
| Energy contribution | -20.62 |
| Covariance contribution | 0.57 |
| Combinations/Pair | 1.64 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.792130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
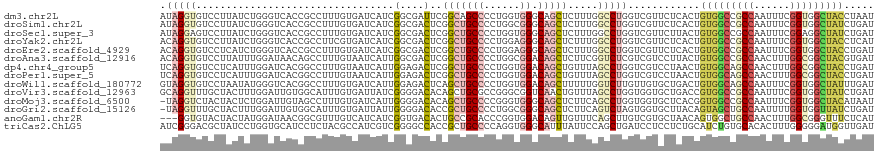
>dm3.chr2L 6862443 119 - 23011544 AUAGGUGUCCUUAUCUGGGUCACCGCCUUUGUGAUCAUCGGCGAUUCGGCAGCCCCUGGUGGGCAGCUCUUUGGCCUGGUCGUUCUCACUGUGGCCGCCAAUUUCGGUGGCUACCUAAU ...((((.......(..(((((.((((..((....))..))))....(((.((((.....)))).)))...)))))..).......))))(((((((((......)))))))))..... ( -47.74, z-score = -2.21, R) >droSim1.chr2L 6662812 119 - 22036055 AUAGGUGUCCUUAUCUGGGUCACCGCCUUUGUGAUCAUCGGCGACUCGGCUGCCCCUGGCGGGCAGCUCUUUGGCCUGGUCGUUCUCACUGUGGCCGCCAAUUUCGGUGGCUAUCUGAU ...((((.(((.....))).))))........(((((..(((.(...((((((((.....))))))))...).))))))))....(((..(((((((((......))))))))).))). ( -51.40, z-score = -3.00, R) >droSec1.super_3 2402592 119 - 7220098 AUAGGAGUCCUUAUCUGGGUCACCGCCUUUGUGAUCAUCGGCGACUCGGCUGCCCCUGGUGGGCAGCUCUUUGGCCUGGUCGUUCUUACUGUGGCCGCCAAUUUCGGAGGCUAUCUGAU .((((((.(.....(..(((((.((((..((....))..))))....((((((((.....))))))))...)))))..)..)))))))..((((((.((......)).))))))..... ( -45.00, z-score = -1.06, R) >droYak2.chr2L 16282467 119 + 22324452 ACAGGUGUCCUUAUCUGGGUCACCGCCUUCGUGAUCAUCGGCGACUCGGCUGCCCCUGGAGGGCAGCUCUUUGGCCUGGUCGUUCUCACUGUGGCCGCCAAUUUCGGUGGCUACCUCAU ...((((.(((.....))).))))........(((((..(((.(...((((((((.....))))))))...).)))))))).........(((((((((......)))))))))..... ( -52.10, z-score = -2.94, R) >droEre2.scaffold_4929 15781418 119 - 26641161 ACAGGUGUCCUCAUCUGGGUCACCGCCUUUGUGAUCAUCGGCGACUCGGCUGCCCCUGGAGGGCAGCUCUUUGGCCUGGUCGUUCUCACUGUGGCCGCCAAUUUCGGUGGCUACCUGAU .((((((....(..(..(((((.((((..((....))..))))....((((((((.....))))))))...)))))..)..)....))).(((((((((......)))))))))))).. ( -55.20, z-score = -3.38, R) >droAna3.scaffold_12916 8449787 119 - 16180835 ACAGGUGUCCUUAUUUGGAUAACAGCCUUUGUAAUCAUUGGCGACUCGGCUGCCCCUGGCGGACAGCUCUUCGGUCUCGUCGUCCUUACUGUGGCCGCCAACUUUGGCGGCUACGUGAU .(((((((((......))))).(((((.((((........))))...)))))..))))..((((.((.(....)....)).))))((((.((((((((((....)))))))))))))). ( -48.00, z-score = -3.24, R) >dp4.chr4_group5 359937 119 - 2436548 UCAGGUGUCCUCAUUUGGAUCACGGCCUUUGUAAUCAUUGGAGACUCGGCUGCCCCUGGUGGACAGCUGUUUAGCCUGGUCGUCCUAACUGUGGCAGCCAACUUUGGCGGCUACCUGAU ((((((((((......))))...((((..(((...((..(((((((.(((((..((((.....)))..)..))))).)))).)))....))..)))((((....)))))))))))))). ( -43.10, z-score = -1.12, R) >droPer1.super_5 4857486 119 - 6813705 UCAGGUGUCCUCAUUUGGAUCACGGCCUUUGUAAUCAUUGGAGACUCGGCUGCCCCUGGUGGACAGCUGUUUAGCCUGGUCGUCCUAACUGUGGCAGCCAACUUUGGCGGCUACCUGAU ((((((((((......))))...((((..(((...((..(((((((.(((((..((((.....)))..)..))))).)))).)))....))..)))((((....)))))))))))))). ( -43.10, z-score = -1.12, R) >droWil1.scaffold_180772 2037021 119 + 8906247 GUAGGUGUCCUAAUAUGGGUCACGGCCUUUGUGAUCAUUGGAGACUCAGCUGCCCCUGGUGGACAGCUUUUUGGUCUUGUUGUGCUGACUGUGGCAGCCAAUUUCGGUGGCUAUUUGAU ....(((.(((.....))).))).(((...((.(.(((..((((((.(((((((......)).)))))....))))))...))).).))...)))(((((.......)))))....... ( -36.00, z-score = 0.22, R) >droVir3.scaffold_12963 5400818 119 + 20206255 GCAGGUUUGCUACUUUGGAUUGUGGCAUUUGUGAUUAUCGGGGACACAGCUGCGCCGGGCGGUCAACUGUUUAGCCUGGUGGUGCUGACCGUGGCCGCCAAUUUCGGUGGCUAUCUGAU .((.(..((((((........))))))..).))...(((((((...((((..((((((((.............))))))))..)))).))(((((((((......)))))))))))))) ( -49.82, z-score = -2.45, R) >droMoj3.scaffold_6500 14595858 118 + 32352404 -UAGGUCUACUACUCUGGAUUGUAGCCUUUGUGAUCAUUGGGGACACAGCUGCCCCGGGUGGGCAGCUCUUCAGCCUGGUGGUGCUCACGGUGGCCGCCAAUUUCGGUGGCUACAUAAU -.(((.((((...........))))))).(((((((((..((.....((((((((.....))))))))......))..))))...)))))(((((((((......)))))))))..... ( -50.10, z-score = -2.18, R) >droGri2.scaffold_15126 4560196 118 - 8399593 -UAGGUUUGCUACUUUGGAUUGUGGCAUUUGUGAUUAUUGGGGACACCGCUGCCCCUGGCGGGCAGCUCUUCAGUCUAGUGGUGCUUACAGUAGCUGCCAAUUUUGGUGGUUAUCUGAU -(((((..((((((..((((((.((.............((....))..(((((((.....))))))).)).)))))))))))))))))(((((((..(((....)))..)))).))).. ( -43.90, z-score = -1.99, R) >anoGam1.chr2R 15238025 116 - 62725911 ---GGUGUACUACUAUGGAUAACGGCGUUUGUCAUCAUCGGUGACACUGCCGCACCCGGUGGACAGUUGUUUCAGCUUGUCGUGCUAACAGUGGCUGCCAACUUUGGCGGGUUUCUCAU ---......(((((..((....(((((..((((((.....)))))).)))))...))((((((((((((...)))).)))).))))...)))))((((((....))))))......... ( -35.70, z-score = 0.14, R) >triCas2.ChLG5 17199765 119 - 18847211 AUCGGGACGCUAUCCUGGUGCAUCCUCUACGCCAUCGUCGGGGCCACCGCUGCCCCAGGUGGGCAUUUAUUCCAGCUGAUCCUCCUCUGCAUCUGUGCACACUUUGGGGGAUGGUUGAU (((((((.....)))))))...........((((((...(((((.......))))).))).)))........(((((.(((((((..((((....))))......)))))))))))).. ( -41.20, z-score = 0.42, R) >consensus AUAGGUGUCCUAAUCUGGAUCACCGCCUUUGUGAUCAUCGGCGACUCGGCUGCCCCUGGUGGGCAGCUCUUUGGCCUGGUCGUGCUCACUGUGGCCGCCAAUUUCGGUGGCUACCUGAU .(((((.................................(....)..(((((((......)).))))).....)))))............(((((((((......)))))))))..... (-20.06 = -20.62 + 0.57)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:22:22 2011