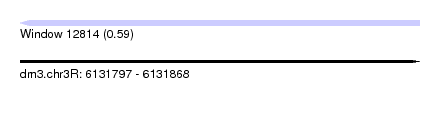
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,131,797 – 6,131,868 |
| Length | 71 |
| Max. P | 0.589939 |

| Location | 6,131,797 – 6,131,868 |
|---|---|
| Length | 71 |
| Sequences | 10 |
| Columns | 77 |
| Reading direction | reverse |
| Mean pairwise identity | 62.40 |
| Shannon entropy | 0.74900 |
| G+C content | 0.61759 |
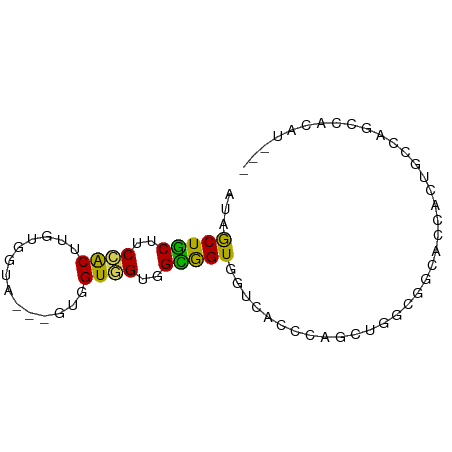
| Mean single sequence MFE | -28.82 |
| Consensus MFE | -11.16 |
| Energy contribution | -10.35 |
| Covariance contribution | -0.81 |
| Combinations/Pair | 1.67 |
| Mean z-score | -0.85 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.589939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
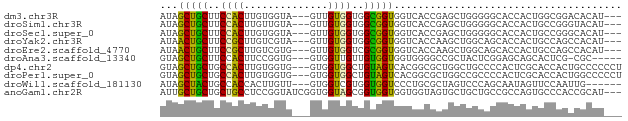
>dm3.chr3R 6131797 71 - 27905053 AUAGCUGCUUCCACUUGUGGUA---GUUGUGGUGGCGGUGGUCACCGAGCUGGGGGCACCACUGGCGGACACAU--- ..(((((((.........))))---)))(((.((.(((((((..((.......))..))))))).))..)))..--- ( -24.70, z-score = 0.46, R) >droSim1.chr3R 12261533 71 + 27517382 AUAGCUGCUUCCACUUGUUGUA---GUUGUGGUGGCGGUGGUCACCGAGCUGGGGGCACCACUGCCGGGUACAU--- ((((((((...........)))---)))))..((((((((((..((.......))..)))))))))).......--- ( -26.50, z-score = -0.19, R) >droSec1.super_0 5320216 71 + 21120651 AUAGCUGCUUCCACUUGUGGUA---GUUGUGGUGGCGGUGGUCACCGAGCUGGGGGCACCACUGCCGGGCACAU--- ..(((((((.........))))---)))(((.((((((((((..((.......))..))))))))).).)))..--- ( -28.10, z-score = -0.09, R) >droYak2.chr3R 10168140 71 - 28832112 AUAACUGCUUCCGCUUGUCGUA---GUUGUGGUGGCGGUGGUCACCAAGCUGGCAGCACCACUGCCAGCCACAU--- ((((((((...........)))---)))))((((((....))))))..((((((((.....)))))))).....--- ( -30.40, z-score = -2.14, R) >droEre2.scaffold_4770 2271449 71 + 17746568 AUAACUGCUUCCGCUUGUCGUG---GUUGUGGUCGCGGUGGUCACCAAGCUGGCAGCACCACUGCCAGCCACAU--- ...((..(..((((.....)))---)..)..))...(((....)))..((((((((.....)))))))).....--- ( -27.00, z-score = -1.06, R) >droAna3.scaffold_13340 22742263 68 + 23697760 GUAGCUGCUUCCACUUCCGGUG---GUGGUUGUUGUGGUGGUGGGGCCGCUACUCGGAGCAGCACUCG-CGC----- ...((((((((((((......)---)))......(((((((.....)))))))..)))))))).....-...----- ( -28.00, z-score = -0.80, R) >dp4.chr2 23461077 74 + 30794189 GUAGCUGCUGCCACUUGUGGUG---GUGGUGGCUGUAGUCACGGCGCUGGCUGCCCCACUCGCACCACUGCCCCCCU .((((..(..((((.....)))---)..)..)))).......((((.(((.(((.......)))))).))))..... ( -31.20, z-score = -0.27, R) >droPer1.super_0 10626848 74 + 11822988 GUAGCUGCUGCCACUUGUGGUG---GUGGUGGCUGUAGUCACGGCGCUGGCCGCCCCACUCGCACCACUGGCCCCCU .........((((..(((((((---(.(((((((...((....))...))))))))))).))))....))))..... ( -30.80, z-score = 0.37, R) >droWil1.scaffold_181130 1686301 68 + 16660200 AUAGCUACUGCCACCACUUGUU---GUGGUCGUGGUGGUCCCUGCGCUAGUCCCAGCAAUAGUUCCAAUUG------ ...(((((..(.(((((.....---))))).)..)))))......(((......)))..............------ ( -19.60, z-score = -1.40, R) >anoGam1.chr2R 33561265 74 - 62725911 AUUGCUGCUGCUGCCUCCGGUAUCGGUGGUAGCGGUGGUGGUGGUAGUGCUGCUGCCGCCAGUGCCCACCGCAU--- ..(((....((((((.(((....))).))))))((((((((((((((.....)))))))))....)))))))).--- ( -41.90, z-score = -3.42, R) >consensus AUAGCUGCUUCCACUUGUGGUA___GUGGUGGUGGCGGUGGUCACCCAGCUGGCGGCACCACUGCCAGCCACAU___ ...(((((..((((..............))))..)))))...................................... (-11.16 = -10.35 + -0.81)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:02:38 2011