
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,050,787 – 6,050,888 |
| Length | 101 |
| Max. P | 0.571426 |

| Location | 6,050,787 – 6,050,888 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 59.57 |
| Shannon entropy | 0.90763 |
| G+C content | 0.40685 |
| Mean single sequence MFE | -21.54 |
| Consensus MFE | -6.85 |
| Energy contribution | -6.30 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.33 |
| Mean z-score | -0.86 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.571426 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6050787 101 + 27905053 AUGAGAAUUUGCACAAGCGGGAGGAUAUCAAGUGUCCCUGUGAGUAACAGGUUUCUAGUUUCG-UAUGAAUUCUAUAAUGUU-AAAAAUGCUAAAUAUUUUCU-- ...(((((((..((((((.(((((((((...))))))((((.....))))...))).)))).)-)..)))))))........-....................-- ( -20.00, z-score = -0.64, R) >droSim1.chr3R 12188780 100 - 27517382 AUGCGAAUUUGCACAAGCGGGAGGACAUCAAGUGUCCCUGUGAGUAAAAGGUUUCUAAUUUCG-CAGGAAUUCUAUAAUGUU-AAA-UUGCUGAAUAUUUUCU-- .((((....))))..(((((...(((((..((.((.((((((((....((....))...))))-)))).)).))...)))))-...-)))))...........-- ( -21.60, z-score = -0.46, R) >droSec1.super_0 5250180 99 - 21120651 AUGAGAAUUUGCACAAGCGGGAGGACAUCAAGUGUCCCUGUGAGUAAAAGGUUUCUAGUUUCG-CAGGAAUUCUAUAAUGUU-AA--UUGCUGCACAUUCUCU-- ..((((((.((((...((((...(((((..((.((.((((((((....((....))...))))-)))).)).))...)))))-..--)))))))).)))))).-- ( -29.10, z-score = -2.31, R) >droYak2.chr3R 10095639 103 + 28832112 AUGAGAAUUUGCACAAGCGUGAGGACAUCAAGUGUCCCUGUGAGUAAAAGGUUUCUAAUUUUCGCAGGCAUACUGUAAUUUU-AAA-AUGUUGUAUAUAUUUUCU ..((((((.((.((((.(((.((((.....(((((.((((((((....((....))....)))))))).)))))....))))-...-))))))).)).)))))). ( -23.70, z-score = -1.57, R) >droEre2.scaffold_4770 2198872 103 - 17746568 AUGAGAAUUUGCACAAGCGGGAGGACAUCAAGUGUCCCUGUGAGUAGAGGGUUUCUGGUUUUGGCAGGAACACUAUAAUUUC-AGA-AUGCUGUACAUAUUUUCC ..((((((.((.(((.(((...((((((...))))))(((.((....((.(((((((.......))))))).))....)).)-)).-.)))))).)).)))))). ( -27.80, z-score = -1.06, R) >droAna3.scaffold_13340 22677536 96 - 23697760 ACCAGAAUCUUCACAAACGCGAGGACAUCAAGUGCCCCUGUGAGUUGCU-GUCUUGCCGUCAUGUCCUACCUGCUAAUCCUGCUAAUUUGAUCCUCU-------- ....................(((((..((((((......((((...((.-.....))..)))).........((.......))..))))))))))).-------- ( -15.20, z-score = 0.61, R) >dp4.chr2 23354369 96 - 30794189 ACGAGAAUCUACAUAAGCGCGAGGACAUCAAGUGUCCCUGUAAGUAUCC--UCCUCCUUUGCCAGCAAAGAGGUAGCCUUCAGAGACAUUUUCCUUCU------- ....................(((((....(((((((.(((.(((...(.--.((((.((((....))))))))..).)))))).))))))))))))..------- ( -23.80, z-score = -1.52, R) >droPer1.super_0 10542355 96 - 11822988 ACGAGAAUCUACAUAAGCGCGAGGAUAUCAAGUGUCCCUGUAAGUAUGC--UCCUCCUUUGCCAGCAAAGAGGUAGCCUUCAGCGACGUUUUCCUUCU------- ...((((.......(((((((.((((((...))))))(((.(((...((--(((((.((((....)))))))).))))))))))).)))))...))))------- ( -23.70, z-score = -0.86, R) >droWil1.scaffold_181108 940917 93 + 4707319 AUGAGAAUCUACACAAGCGUGAGGAUAUUAAAUGUCCCUGUAAGUA--UAAA--GAGAUCACC-UAAGAUUUGUAAGAUGUUUUGAUCUUUUGUUUAU------- ...........(((....))).((((((...))))))......(((--..((--(((((((..-.((.(((.....))).)).)))))))))...)))------- ( -14.60, z-score = -0.07, R) >droVir3.scaffold_13047 18139119 98 - 19223366 AUGAGAAUCUGCACAAGCGUGAGGACAUCAAAUGUCCCUGUAAGUAGCCAAA--CAUAUUGAU-UGCAACGCGUGCAUUC----AAUCUAAUCUCUACGCUCGGC ..........((.(.((((((.((((((...))))))...............--...(((((.-((((.....)))).))----)))........)))))).))) ( -22.00, z-score = -0.27, R) >droMoj3.scaffold_6540 29043113 94 - 34148556 AUGAGAAUCUGCACAAGCGCGAGGAUAUCAAAUGCCCCUGUAAGUGAUCAGCCUCUACUUCUCGAUCUACAUGGAAAUUAUUCUCG--UAAUCCCU--------- ..(((((...((......))((((..((((..(((....)))..))))...))))...)))))(((.(((..((((....)))).)--)))))...--------- ( -18.80, z-score = -0.52, R) >droGri2.scaffold_14906 5192705 100 + 14172833 AUGAGAAUCUGCACAAACGCGAGGACAUUAAAUGUCCCUGUAAGUAAACCAC--AAAAUUAGUGCACUUUACCAUUAUUACAGAAGUUCAUUAACCACGUUC--- (((((..((((.......(((.((((((...)))))).)))..(((((.(((--.......)))...)))))........))))..)))))...........--- ( -18.20, z-score = -1.60, R) >consensus AUGAGAAUCUGCACAAGCGCGAGGACAUCAAGUGUCCCUGUAAGUAAACGGUUUCUAAUUUCC_CACGAAUUCUAUAAUGUU_AAA_AUGUUCCAUAU__U____ ............((..(((.(.((((((...))))))))))..))............................................................ ( -6.85 = -6.30 + -0.55)
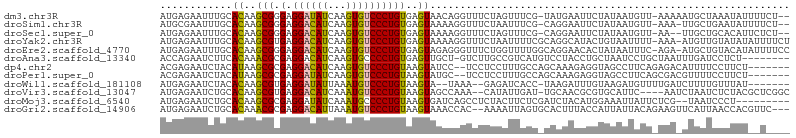


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:02:31 2011