| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,021,429 – 6,021,496 |
| Length | 67 |
| Max. P | 0.625429 |

| Location | 6,021,429 – 6,021,496 |
|---|---|
| Length | 67 |
| Sequences | 14 |
| Columns | 69 |
| Reading direction | forward |
| Mean pairwise identity | 83.40 |
| Shannon entropy | 0.39063 |
| G+C content | 0.54229 |
| Mean single sequence MFE | -22.16 |
| Consensus MFE | -13.54 |
| Energy contribution | -13.69 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.625429 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
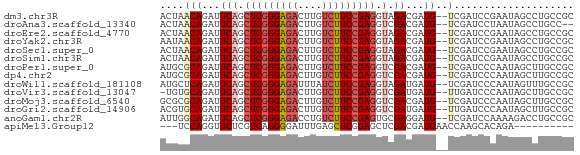
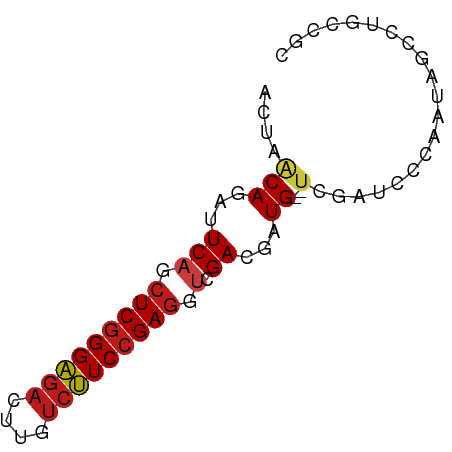
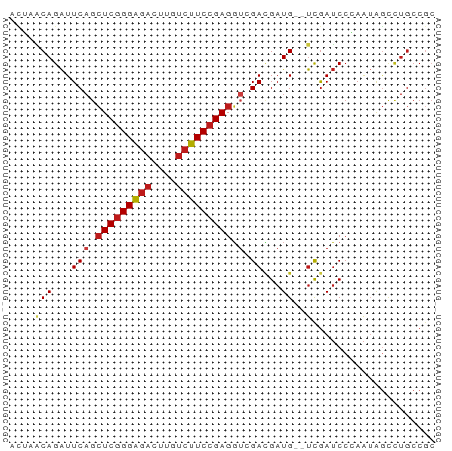
>dm3.chr3R 6021429 67 + 27905053 ACUAACAGAUUCAGCUCGGGAGACUUGUCUUCCGAGGUAGACGAUG--UCGAUCCGAAUAGCCUGCCGC .............((((((((((....))))))))(((((.(....--(((...)))...).))))))) ( -20.50, z-score = -1.09, R) >droAna3.scaffold_13340 22656375 65 - 23697760 ACUAACAGAUUCAGCUCGGGAGACUUGUCUUCCGAGGUCGACGAUG--UCGAUCCUAAUAGCCUGCC-- .....(((......(((((((((....)))))))))((((((...)--))))).........)))..-- ( -21.90, z-score = -2.04, R) >droEre2.scaffold_4770 2175008 67 - 17746568 ACUAACAGAUUCAGCUCGGGAGACUUGUCUUCCGAGGUAGACGAUG--UCGAUCCGAAUAGCCUGCCGC .............((((((((((....))))))))(((((.(....--(((...)))...).))))))) ( -20.50, z-score = -1.09, R) >droYak2.chr3R 10071547 67 + 28832112 AAUAACAGAUUCAGCUCGGGAGACUUGUCUUCCGAGGUAGACGAUG--UCGAUCCGAAUAGCCUGCCGC .............((((((((((....))))))))(((((.(....--(((...)))...).))))))) ( -20.50, z-score = -1.31, R) >droSec1.super_0 5226547 67 - 21120651 ACUAACAGAUUCAGCUCGGGAGACUUGUCUUCCGAGGUAGACGAUG--UCGAUCCGAAUAGCCUGCCGC .............((((((((((....))))))))(((((.(....--(((...)))...).))))))) ( -20.50, z-score = -1.09, R) >droSim1.chr3R 12164049 67 - 27517382 ACUAACAGAUUCAGCUCGGGAGACUUGUCUUCCGAGGUAGACGAUG--UCGAUCCGAAUAGCCUGCCGC .............((((((((((....))))))))(((((.(....--(((...)))...).))))))) ( -20.50, z-score = -1.09, R) >droPer1.super_0 10518174 67 - 11822988 AUGCGCAGAUUCAGCUCGGGAGACUUGUCUUCCGAGGUCGACGAUG--UCGAUCCCAAUAGCUUGCCGC ..(((((.....(((((((((((....))))))))(((((((...)--))))))......))))).))) ( -24.10, z-score = -1.52, R) >dp4.chr2 23330386 67 - 30794189 AUGCGCAGAUUCAGCUCGGGAGACUUGUCUUCCGAGGUCGACGAUG--UCGAUCCCAAUAGCUUGCCGC ..(((((.....(((((((((((....))))))))(((((((...)--))))))......))))).))) ( -24.10, z-score = -1.52, R) >droWil1.scaffold_181108 912363 67 + 4707319 AUGCUCAGAUUCAGCUCGGGAGAUUUAUCUUCCGAGGUAGAUGAUG--UCGAUCCCAAUAGUUUGCCGC ..(((.......)))((((((((....))))))))(((((((..((--.......))...))))))).. ( -18.00, z-score = -0.74, R) >droVir3.scaffold_13047 18112228 66 - 19223366 -UGUGCAGAUUCAGCUCGGGAGACUUGUCUUCCGAGGUCGAUGAUG--UUGAUCCCAAUAGCUUGCCGC -.(.(((((((.(.(((((((((....))))))))).).)))..((--(((....)))))..))))).. ( -20.70, z-score = -1.04, R) >droMoj3.scaffold_6540 29012236 67 - 34148556 GCGCGCAGAUUCAGCUCGGGAGACUUGUCUUCCGAGGUCGACGAUG--UCGAUCCCAAUAGCUUGCCGC (((.(((.....(((((((((((....))))))))(((((((...)--))))))......))))))))) ( -27.40, z-score = -2.20, R) >droGri2.scaffold_14906 5163574 67 + 14172833 ACGUGCAGAUUCAGCUCGGGAGACUUGUCUUCCGAGGUCGAUGAUG--UUGAUCCCAAUAGCUUGCCGC .((.(((((((.(.(((((((((....))))))))).).)))..((--(((....)))))..)))))). ( -22.30, z-score = -1.46, R) >anoGam1.chr2R 36296710 67 + 62725911 AUUGGCAGAUUCAGCUCGGGAGACCUGUCUUCCGAGUGCGAGGAUG--UCGAUCCAAAAGACCUGCCGC ...(((((..((.((((((((((....))))))))))..))((((.--...)))).......))))).. ( -30.60, z-score = -3.74, R) >apiMel3.Group12 4200104 56 - 9182753 ---UCCAGGUUCUCGUCAGGGGAUUUGAGCUCGGAGCUCGACGAUGAACCAAGCACAGA---------- ---....((((((((((.(((..((((....)))).)))))))).))))).........---------- ( -18.70, z-score = -1.31, R) >consensus ACUAACAGAUUCAGCUCGGGAGACUUGUCUUCCGAGGUCGACGAUG__UCGAUCCCAAUAGCCUGCCGC ..............(((((((((....)))))))))................................. (-13.54 = -13.69 + 0.15)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:02:27 2011