
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,017,716 – 6,017,808 |
| Length | 92 |
| Max. P | 0.989315 |

| Location | 6,017,716 – 6,017,808 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 69.65 |
| Shannon entropy | 0.58030 |
| G+C content | 0.49135 |
| Mean single sequence MFE | -29.41 |
| Consensus MFE | -13.60 |
| Energy contribution | -13.74 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.939265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
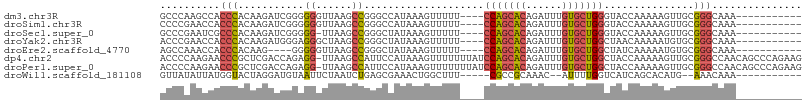
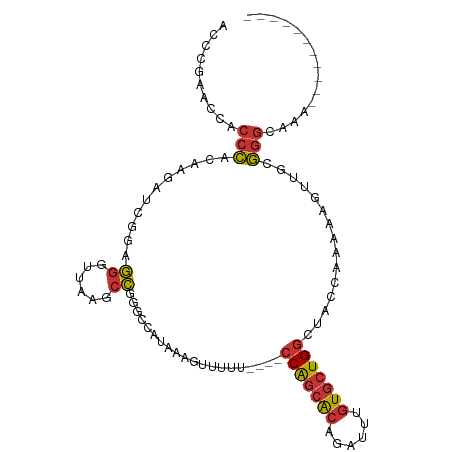
>dm3.chr3R 6017716 92 + 27905053 GCCCAAGCCACCCACAAGAUCGGGGGGUUAAGCCGGGCCAUAAAGUUUUU----CCAGCACAGAUUUGUGCUGGGUACCAAAAAGUUGCGGGCAAA----------- ((((..(((.(((........))).((.....)).)))......((...(----(((((((......)))))))).))...........))))...----------- ( -33.40, z-score = -1.43, R) >droSim1.chr3R 12160365 92 - 27517382 CCCCGAACCACCCACAAGAUCGGGGGGUUAAGCCGGGCCAUAAAGUUUUU----CCAGCACAGAUUUGUGCUGGGUACCAAAAAGUUGCGGGCAAA----------- ..(((((((.(((........))).))))....)))(((...((.(((((----(((((((......)))))))).....)))).))...)))...----------- ( -31.70, z-score = -1.08, R) >droSec1.super_0 5222950 91 - 21120651 GCCCGAAUCGCCCACAAGAUCGGGGG-UUAAGCCGGGCUAUAAAGUUUUU----CCAGCACAGAUUUGUGCUGGGUACCAAAAAGUUGCGGGCAAA----------- (((((...(((((.(......)..((-.....))))))......((...(----(((((((......)))))))).))......)...)))))...----------- ( -33.20, z-score = -1.97, R) >droYak2.chr3R 10067901 92 + 28832112 ACCCGAACCACCCACAAGAUGGGAGGGCUAAGCCGGGCUAUAAAGUUUUU----CCAGCACAGAUUUGUGCUGGCUAACAAAAAUGUGCGGGCAAA----------- .((((..((.((((.....))))..(((...)))))........(((...----(((((((......)))))))..))).........))))....----------- ( -30.90, z-score = -1.58, R) >droEre2.scaffold_4770 2171433 88 - 17746568 AGCCAAACCACCCACAAG----GGGGGUUAAGCCGGGCUAUAAAGUUUUU----CCAGCACAGAUUUGUGCUGGCUAUCAAAAAUGUGCGGGCAAA----------- .(((.((((.(((....)----)).))))....((.((............----(((((((......)))))))...........)).)))))...----------- ( -30.60, z-score = -1.58, R) >dp4.chr2 23326797 106 - 30794189 ACCCCAAGAACCCGCUCGACCAGAGG-UUAAGCCAUUCCAUAAAGUUUUUUUAUCCAGCACAGAUUUGUGCUGGCUACCAAAAAGUUGCGGGCCAACAGCCCAGAAG .............(((..(((...))-)..)))..(((..(((..((((..((.(((((((......))))))).))..))))..))).((((.....)))).))). ( -28.50, z-score = -2.68, R) >droPer1.super_0 10514623 106 - 11822988 ACCCCAAGAACCCGCUCGACCAGAGG-UUAAGCCAUUCCAUAAAGUUUUUUUAUCCAGCACAGAUUUGUGCUGGCUACCAAAAAGUUGCGGGCCAACAGCCCAGAAG .............(((..(((...))-)..)))..(((..(((..((((..((.(((((((......))))))).))..))))..))).((((.....)))).))). ( -28.50, z-score = -2.68, R) >droWil1.scaffold_181108 907768 87 + 4707319 GUUAUAUUAUGGUACUAGGAUGUAAUUCUAAUCUGAGCGAAACUGGCUUU-----CGCCGCAAAC--AUUUUGGUCAUCAGCACAUG--AAACAAA----------- (((.(((..((((((((((((((...((......))((((((.....)))-----))).....))--)))))))).))))....)))--.)))...----------- ( -18.50, z-score = -1.11, R) >consensus ACCCGAACCACCCACAAGAUCGGAGGGUUAAGCCGGGCCAUAAAGUUUUU____CCAGCACAGAUUUGUGCUGGCUACCAAAAAGUUGCGGGCAAA___________ ..........(((...........((......))...........(((((....(((((((......))))))).....))))).....)))............... (-13.60 = -13.74 + 0.14)
| Location | 6,017,716 – 6,017,808 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 69.65 |
| Shannon entropy | 0.58030 |
| G+C content | 0.49135 |
| Mean single sequence MFE | -30.39 |
| Consensus MFE | -19.11 |
| Energy contribution | -19.68 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.989315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
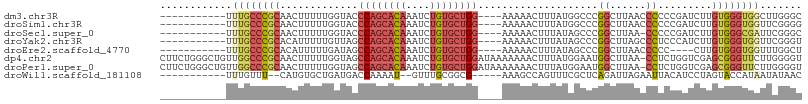
>dm3.chr3R 6017716 92 - 27905053 -----------UUUGCCCGCAACUUUUUGGUACCCAGCACAAAUCUGUGCUGG----AAAAACUUUAUGGCCCGGCUUAACCCCCCGAUCUUGUGGGUGGCUUGGGC -----------...(((....(((....)))..((((((((....))))))))----...........)))...((((((.((((((......)))).)).)))))) ( -29.80, z-score = -0.69, R) >droSim1.chr3R 12160365 92 + 27517382 -----------UUUGCCCGCAACUUUUUGGUACCCAGCACAAAUCUGUGCUGG----AAAAACUUUAUGGCCCGGCUUAACCCCCCGAUCUUGUGGGUGGUUCGGGG -----------...(((.((.(((....)))..((((((((....))))))))----............))..)))......((((((.((.......)).)))))) ( -30.50, z-score = -0.83, R) >droSec1.super_0 5222950 91 + 21120651 -----------UUUGCCCGCAACUUUUUGGUACCCAGCACAAAUCUGUGCUGG----AAAAACUUUAUAGCCCGGCUUAA-CCCCCGAUCUUGUGGGCGAUUCGGGC -----------.((((((((((....((((...((((((((....))))))))----................((.....-)).))))..))))))))))....... ( -32.00, z-score = -2.62, R) >droYak2.chr3R 10067901 92 - 28832112 -----------UUUGCCCGCACAUUUUUGUUAGCCAGCACAAAUCUGUGCUGG----AAAAACUUUAUAGCCCGGCUUAGCCCUCCCAUCUUGUGGGUGGUUCGGGU -----------...(((((.((......(((..((((((((....))))))))----...)))..........(((...)))(.(((((...))))).))).))))) ( -31.50, z-score = -1.91, R) >droEre2.scaffold_4770 2171433 88 + 17746568 -----------UUUGCCCGCACAUUUUUGAUAGCCAGCACAAAUCUGUGCUGG----AAAAACUUUAUAGCCCGGCUUAACCCCC----CUUGUGGGUGGUUUGGCU -----------.......((.((....))....((((((((....))))))))----............))..((((.((((.((----(....))).)))).)))) ( -29.10, z-score = -1.83, R) >dp4.chr2 23326797 106 + 30794189 CUUCUGGGCUGUUGGCCCGCAACUUUUUGGUAGCCAGCACAAAUCUGUGCUGGAUAAAAAAACUUUAUGGAAUGGCUUAA-CCUCUGGUCGAGCGGGUUCUUGGGGU .....((((.....)))).....((((((....((((((((....)))))))).)))))).((((((.(((.(.(((..(-((...)))..))).).))).)))))) ( -36.20, z-score = -1.92, R) >droPer1.super_0 10514623 106 + 11822988 CUUCUGGGCUGUUGGCCCGCAACUUUUUGGUAGCCAGCACAAAUCUGUGCUGGAUAAAAAAACUUUAUGGAAUGGCUUAA-CCUCUGGUCGAGCGGGUUCUUGGGGU .....((((.....)))).....((((((....((((((((....)))))))).)))))).((((((.(((.(.(((..(-((...)))..))).).))).)))))) ( -36.20, z-score = -1.92, R) >droWil1.scaffold_181108 907768 87 - 4707319 -----------UUUGUUU--CAUGUGCUGAUGACCAAAAU--GUUUGCGGCG-----AAAGCCAGUUUCGCUCAGAUUAGAAUUACAUCCUAGUACCAUAAUAUAAC -----------..((((.--...(((((((((........--(((((.((((-----(((.....))))))))))))........))))..)))))...)))).... ( -17.79, z-score = -1.24, R) >consensus ___________UUUGCCCGCAACUUUUUGGUAGCCAGCACAAAUCUGUGCUGG____AAAAACUUUAUGGCCCGGCUUAACCCCCCGAUCUUGUGGGUGGUUCGGGU ............((((((((...(((((.....((((((((....))))))))....)))))...........((......)).........))))))))....... (-19.11 = -19.68 + 0.56)
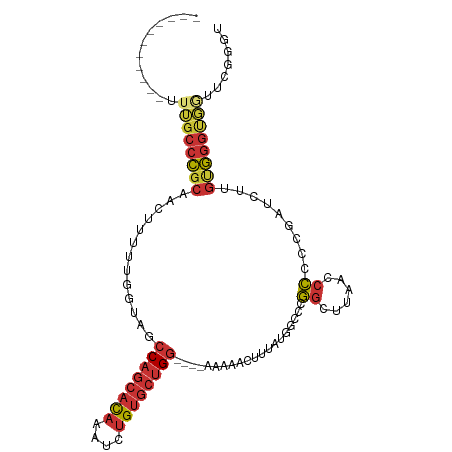
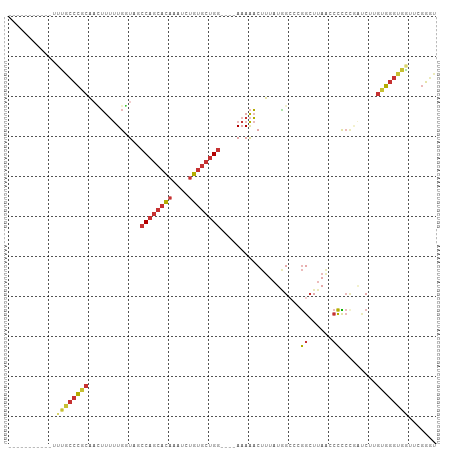
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:02:26 2011