
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 5,984,455 – 5,984,535 |
| Length | 80 |
| Max. P | 0.997602 |

| Location | 5,984,455 – 5,984,535 |
|---|---|
| Length | 80 |
| Sequences | 6 |
| Columns | 82 |
| Reading direction | forward |
| Mean pairwise identity | 70.50 |
| Shannon entropy | 0.55681 |
| G+C content | 0.40833 |
| Mean single sequence MFE | -21.95 |
| Consensus MFE | -6.71 |
| Energy contribution | -8.47 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.884136 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
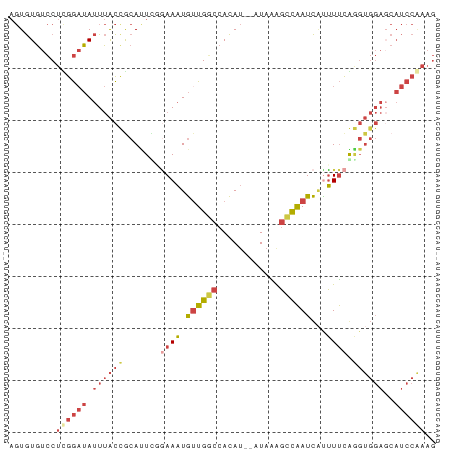
>dm3.chr3R 5984455 80 + 27905053 AGUGUGUCCUCGGAUAUUUACCGCAUUUGAAAAUGUUGGCCACAU--AUAAAGCCAAUCAUUUUCGGGUGGAUCAUCCAAAG ...........(((((((((((.....(((((((((((((.....--.....))))).))))))))))))))).)))).... ( -23.50, z-score = -2.72, R) >droVir3.scaffold_12855 5320536 64 - 10161210 --UAUUUACGCAACGAAUAGUCACAAUCCCAACUAUUGUCUAUAAUAAUAGAUGUGAUUGUGUCUA---------------- --...............(((.(((((((.((.(((((((.....))))))).)).))))))).)))---------------- ( -17.60, z-score = -4.38, R) >droSim1.chr3R 12126312 80 - 27517382 AGUGUGUCCUUGGAUAUUUACCGCACUCGGAAAUGUUGGCCACAU--AUAGAGCCAAUCAUUUUCGGGUGGAGCAUCCAAAG .........((((((.(((....((((((((((((((((((....--...).))))).))))))))))))))).)))))).. ( -27.60, z-score = -3.12, R) >droSec1.super_0 5190180 80 - 21120651 AGUGUGUCCUUGGAUAUUUACCACACUUGGAAAUGAUGGCCACAU--AUAGGGCCAAUCAUUUUCGGGUGGAGCAUCCAAAG .........((((((.(((....((((((((((((((((((....--....)))).))))))))))))))))).)))))).. ( -30.00, z-score = -3.42, R) >droYak2.chr3R 10033084 80 + 28832112 AGUGUGUCCUCGGAUAUUUACUGCAUUCUGAAAUGUUGGCCACAU--AUAAAGCCAAUCCCUUUCAAGUGGAGCAUCCGAAG .........((((((.((((((......(((((.((((((.....--.....))))))...)))))))))))..)))))).. ( -19.40, z-score = -1.49, R) >droEre2.scaffold_4770 2138534 80 - 17746568 AGUGUGUCCUUGGAUAUUUACUGCAUUUUGAAAUGUUGUCCACAU--AUAAAGCCAAUCACUUUCAAGUGGAGCAUCCAAAG .........((((((.....(.(((((....))))).)(((((..--..((((.......))))...)))))..)))))).. ( -13.60, z-score = 0.08, R) >consensus AGUGUGUCCUCGGAUAUUUACCGCAUUCGGAAAUGUUGGCCACAU__AUAAAGCCAAUCAUUUUCAGGUGGAGCAUCCAAAG .........((((((.((((((.......((((.((((((............))))))...)))).))))))..)))))).. ( -6.71 = -8.47 + 1.75)


| Location | 5,984,455 – 5,984,535 |
|---|---|
| Length | 80 |
| Sequences | 6 |
| Columns | 82 |
| Reading direction | reverse |
| Mean pairwise identity | 70.50 |
| Shannon entropy | 0.55681 |
| G+C content | 0.40833 |
| Mean single sequence MFE | -23.12 |
| Consensus MFE | -10.24 |
| Energy contribution | -10.63 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.54 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.14 |
| SVM RNA-class probability | 0.997602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
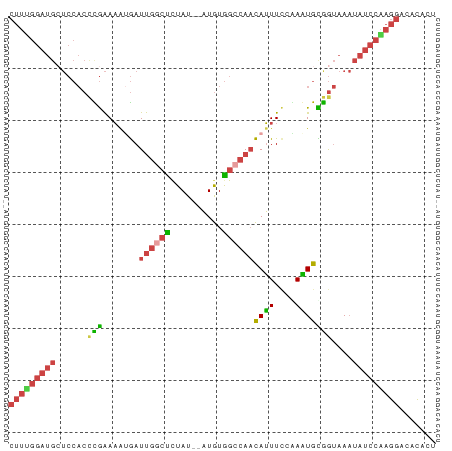
>dm3.chr3R 5984455 80 - 27905053 CUUUGGAUGAUCCACCCGAAAAUGAUUGGCUUUAU--AUGUGGCCAACAUUUUCAAAUGCGGUAAAUAUCCGAGGACACACU (((((((((..((.(..(((((((.((((((....--....)))))))))))))....).))....)))))))))....... ( -25.70, z-score = -3.76, R) >droVir3.scaffold_12855 5320536 64 + 10161210 ----------------UAGACACAAUCACAUCUAUUAUUAUAGACAAUAGUUGGGAUUGUGACUAUUCGUUGCGUAAAUA-- ----------------(((.(((((((.((.(((((.........))))).)).))))))).)))...............-- ( -13.50, z-score = -1.92, R) >droSim1.chr3R 12126312 80 + 27517382 CUUUGGAUGCUCCACCCGAAAAUGAUUGGCUCUAU--AUGUGGCCAACAUUUCCGAGUGCGGUAAAUAUCCAAGGACACACU (((((((((((.(((.((.(((((.((((((....--....))))))))))).)).))).)))....))))))))....... ( -27.40, z-score = -3.69, R) >droSec1.super_0 5190180 80 + 21120651 CUUUGGAUGCUCCACCCGAAAAUGAUUGGCCCUAU--AUGUGGCCAUCAUUUCCAAGUGUGGUAAAUAUCCAAGGACACACU (((((((((..((((.(..(((((((.(((((...--..).)))))))))))....).))))....)))))))))....... ( -28.80, z-score = -3.85, R) >droYak2.chr3R 10033084 80 - 28832112 CUUCGGAUGCUCCACUUGAAAGGGAUUGGCUUUAU--AUGUGGCCAACAUUUCAGAAUGCAGUAAAUAUCCGAGGACACACU (((((((((((.((.((((((..(.((((((....--....))))))).))))))..)).)))....))))))))....... ( -23.70, z-score = -2.55, R) >droEre2.scaffold_4770 2138534 80 + 17746568 CUUUGGAUGCUCCACUUGAAAGUGAUUGGCUUUAU--AUGUGGACAACAUUUCAAAAUGCAGUAAAUAUCCAAGGACACACU (((((((((.(((((...(((((.....)))))..--..)))))...((((....)))).......)))))))))....... ( -19.60, z-score = -1.97, R) >consensus CUUUGGAUGCUCCACCCGAAAAUGAUUGGCUCUAU__AUGUGGCCAACAUUUCCAAAUGCGGUAAAUAUCCAAGGACACACU (((((((((......(((.......((((((.((....)).))))))((((....)))))))....)))))))))....... (-10.24 = -10.63 + 0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:02:23 2011