
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 6,851,951 – 6,852,115 |
| Length | 164 |
| Max. P | 0.988238 |

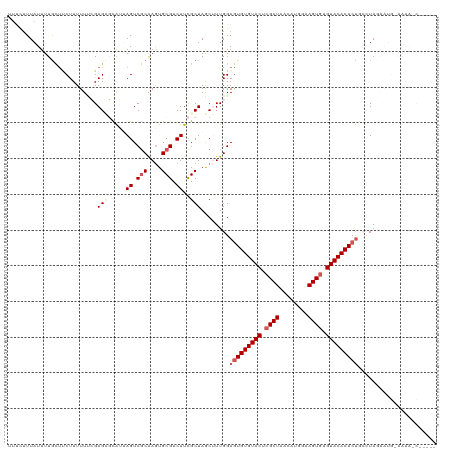
| Location | 6,851,951 – 6,852,071 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.20 |
| Shannon entropy | 0.45492 |
| G+C content | 0.39820 |
| Mean single sequence MFE | -31.74 |
| Consensus MFE | -19.54 |
| Energy contribution | -20.86 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.988238 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 6851951 120 + 23011544 AUUCUUUAUUUUACUUCUUUUUUCGAGAGCAUUAGUCGCAAGUGCGUCUUCGCUAGCUUUGGAAUGUGUACUCGCUAAUAUGGAAGUAUGUACACACUGCAUAGAAUGAAAAAACGCUAU ....(((((((((........(((.(((((...((.(((....))).))......))))).)))(((((((..(((........)))..))))))).....))))))))).......... ( -28.10, z-score = -0.90, R) >droEre2.scaffold_4929 15770796 115 + 26641161 ---UAUUAUUUUGCCACUUUUUCAGAGCUCA--AGUCGCAAGUGCGUCUUUGCUAGCAUUGGAGUGUGUACUCGCUAACUCGAAAGUGUGUACACACUGCCUAGAAUGUAAAAUCUUGUA ---....(((((((...........(((..(--((.(((....))).))).)))......(((((((((((.((((........)))).))))))))).))......)))))))...... ( -33.30, z-score = -2.13, R) >droYak2.chr2L 16271950 118 - 22324452 UUAAUUUAUUUUGCUCCUUUUUGAGAGUUUG--AGUCUCGAAUGCGUCUUUGCUAACGUUGGAGUGUGUACUCGCUAACUGGGCAGUGUGUACACACCGCAUACAAUGUAAAAUCACUUA .......(((((((......((((((.....--..))))))((((((((..((....)).)))((((((((.((((........)))).))))))))))))).....)))))))...... ( -32.80, z-score = -2.04, R) >droSec1.super_3 2392143 104 + 7220098 UUUCUUUAUUUUGCUUCUUUUUUCGAGAGCAUUAGUCGCAAGUGCGUCUUCGCUAACUUUGGAGUGUGUACUCGCUAACAUGAAAGUGUGUACACACUCGCUAU---------------- ...........((((.(((.....)))))))......(((((((((....)))..))))..((((((((((.((((........)))).))))))))))))...---------------- ( -31.90, z-score = -3.13, R) >droSim1.chr2L 6651487 104 + 22036055 UUUCUUUAUUUUGUUUCUUUUUUCGAGCGCAUUAGUCGCAAGUGCGUCUUCGCUAGCUUUGGAGUGUGUACUCGCUAAAAUGGAAGUGUGUACACACUCGCUAU---------------- .........................(((((...((.(((....))).))..))........((((((((((.((((........)))).)))))))))))))..---------------- ( -32.60, z-score = -3.12, R) >consensus UUUCUUUAUUUUGCUUCUUUUUUCGAGAGCAUUAGUCGCAAGUGCGUCUUCGCUAGCUUUGGAGUGUGUACUCGCUAACAUGGAAGUGUGUACACACUGCCUAGAAUG_AAAA_C_____ .....................(((((((.....((.(((....))).))......)).)))))((((((((.((((........)))).))))))))....................... (-19.54 = -20.86 + 1.32)


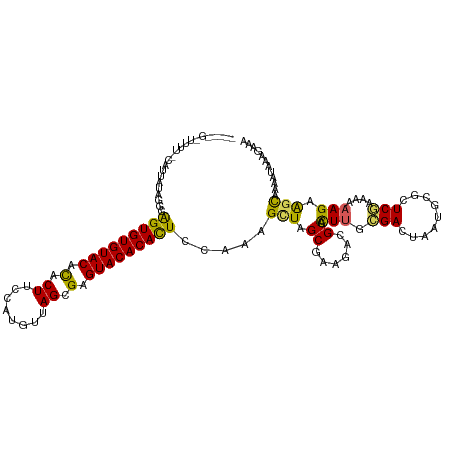
| Location | 6,851,951 – 6,852,071 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.20 |
| Shannon entropy | 0.45492 |
| G+C content | 0.39820 |
| Mean single sequence MFE | -26.66 |
| Consensus MFE | -15.28 |
| Energy contribution | -15.00 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.870533 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 6851951 120 - 23011544 AUAGCGUUUUUUCAUUCUAUGCAGUGUGUACAUACUUCCAUAUUAGCGAGUACACAUUCCAAAGCUAGCGAAGACGCACUUGCGACUAAUGCUCUCGAAAAAAGAAGUAAAAUAAAGAAU ...((((((((.(...((.((.(((((((((...((........))...))))))))).)).))...).))))))))............((((.((.......))))))........... ( -23.70, z-score = -0.60, R) >droEre2.scaffold_4929 15770796 115 - 26641161 UACAAGAUUUUACAUUCUAGGCAGUGUGUACACACUUUCGAGUUAGCGAGUACACACUCCAAUGCUAGCAAAGACGCACUUGCGACU--UGAGCUCUGAAAAAGUGGCAAAAUAAUA--- ....(((........))).((.(((((((((.(.((........)).).)))))))))))..(((((((.(((.(((....))).))--)..)).((.....)))))))........--- ( -28.90, z-score = -1.25, R) >droYak2.chr2L 16271950 118 + 22324452 UAAGUGAUUUUACAUUGUAUGCGGUGUGUACACACUGCCCAGUUAGCGAGUACACACUCCAACGUUAGCAAAGACGCAUUCGAGACU--CAAACUCUCAAAAAGGAGCAAAAUAAAUUAA ....((((((....((((..(((((((....)))))))((.((((((((((....))))....))))))............((((..--.....)))).....)).))))...)))))). ( -25.90, z-score = -1.23, R) >droSec1.super_3 2392143 104 - 7220098 ----------------AUAGCGAGUGUGUACACACUUUCAUGUUAGCGAGUACACACUCCAAAGUUAGCGAAGACGCACUUGCGACUAAUGCUCUCGAAAAAAGAAGCAAAAUAAAGAAA ----------------...((((((((((((.(.((........)).).))))))))))..((((..(((....)))))))))......((((.((.......))))))........... ( -27.30, z-score = -2.32, R) >droSim1.chr2L 6651487 104 - 22036055 ----------------AUAGCGAGUGUGUACACACUUCCAUUUUAGCGAGUACACACUCCAAAGCUAGCGAAGACGCACUUGCGACUAAUGCGCUCGAAAAAAGAAACAAAAUAAAGAAA ----------------.((((((((((((((.(.((........)).).))))))))))....))))(((.((.(((....))).))....))).......................... ( -27.50, z-score = -3.10, R) >consensus _____G_UUUU_CAUUAUAGGCAGUGUGUACACACUUCCAUGUUAGCGAGUACACACUCCAAAGCUAGCGAAGACGCACUUGCGACUAAUGCGCUCGAAAAAAGAAGCAAAAUAAAGAAA ......................(((((((((.(.((........)).).)))))))))................(((....))).................................... (-15.28 = -15.00 + -0.28)

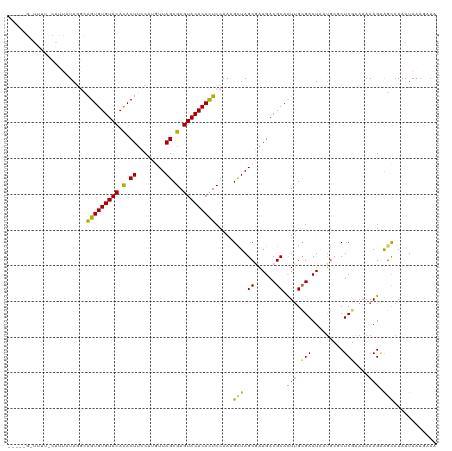
| Location | 6,851,991 – 6,852,111 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.74 |
| Shannon entropy | 0.42224 |
| G+C content | 0.39761 |
| Mean single sequence MFE | -27.83 |
| Consensus MFE | -18.57 |
| Energy contribution | -19.21 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.826103 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 6851991 120 + 23011544 AGUGCGUCUUCGCUAGCUUUGGAAUGUGUACUCGCUAAUAUGGAAGUAUGUACACACUGCAUAGAAUGAAAAAACGCUAUCUUAUUCCAGAACCUCAAAUCUUGUACAGCUUGAACUCUU .(((((...........((((((((((((((..(((........)))..)))))).....((((..(......)..))))...))))))))...........)))))............. ( -22.35, z-score = 0.34, R) >droEre2.scaffold_4929 15770831 119 + 26641161 AGUGCGUCUUUGCUAGCAUUGGAGUGUGUACUCGCUAACUCGAAAGUGUGUACACACUGCCUAGAAUGUAAAAUCUUGUACAUCCUUCAGGAUUGAAAA-UCUGUACAGUUGGAACUUUU .((((..........((((((((((((((((.((((........)))).))))))))).))...)))))..(((((((.........))))))).....-...))))............. ( -32.40, z-score = -1.70, R) >droYak2.chr2L 16271988 119 - 22324452 AAUGCGUCUUUGCUAACGUUGGAGUGUGUACUCGCUAACUGGGCAGUGUGUACACACCGCAUACAAUGUAAAAUCACUUACAUUUUCCAGAACUCCAAA-ACUGUACAGUUUGAAUUUUU .((((((((..((....)).)))((((((((.((((........)))).)))))))))))))..(((((((......))))))).............((-(((....)))))........ ( -30.40, z-score = -1.64, R) >droSec1.super_3 2392183 104 + 7220098 AGUGCGUCUUCGCUAACUUUGGAGUGUGUACUCGCUAACAUGAAAGUGUGUACACACU----------------CGCUAUCAUUUUCCAGAACCCAAAAUCUUGUACAGUUUGAACUUUU ((((((....)))........((((((((((.((((........)))).)))))))))----------------)))).........((((...(((....))).....))))....... ( -25.70, z-score = -1.80, R) >droSim1.chr2L 6651527 104 + 22036055 AGUGCGUCUUCGCUAGCUUUGGAGUGUGUACUCGCUAAAAUGGAAGUGUGUACACACU----------------CGCUAUCAUUUUCCUGAACCUAAAAUGUUGUACAGUUUGAAAUUUU .(((((.......((((....((((((((((.((((........)))).)))))))))----------------))))).((((((.........)))))).)))))............. ( -28.30, z-score = -2.35, R) >consensus AGUGCGUCUUCGCUAGCUUUGGAGUGUGUACUCGCUAACAUGGAAGUGUGUACACACUGC_UA_AAUG_AAAA_CGCUAUCAUUUUCCAGAACCCAAAAUCUUGUACAGUUUGAACUUUU .(((((............(((((((((((((.((((........)))).))))))))............................)))))............)))))............. (-18.57 = -19.21 + 0.64)

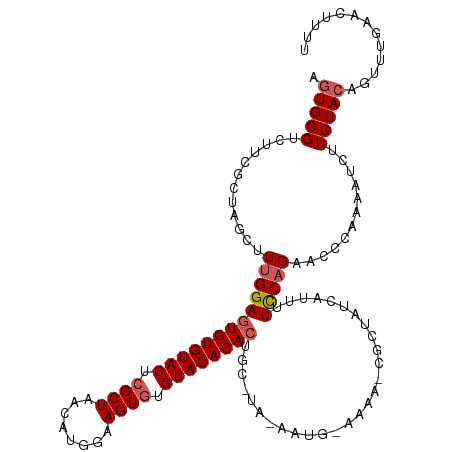
| Location | 6,851,996 – 6,852,115 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.41 |
| Shannon entropy | 0.46253 |
| G+C content | 0.39426 |
| Mean single sequence MFE | -26.16 |
| Consensus MFE | -15.22 |
| Energy contribution | -15.82 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.548077 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 6851996 119 + 23011544 GUCUUCGCUAGCUUUGGAAUGUGUACUCGCUAAUAUGGAAGUAUGUACACACUGCAUAGAAUGAAAAAACGCUAUCUUAUUCCAGAACCUCAAAUCUUGUACAGCUUGAACUCUUUUAG- (..((((..(((((((((((((((((..(((........)))..)))))).....((((..(......)..))))...))))))))....((.....))....)))))))..)......- ( -18.90, z-score = 0.71, R) >droEre2.scaffold_4929 15770836 119 + 26641161 GUCUUUGCUAGCAUUGGAGUGUGUACUCGCUAACUCGAAAGUGUGUACACACUGCCUAGAAUGUAAAAUCUUGUACAUCCUUCAGGAUUGAAAAU-CUGUACAGUUGGAACUUUUCGGAG .......(((((...(((((((((((.((((........)))).))))))))).))((((......(((((((.........))))))).....)-)))....)))))............ ( -32.10, z-score = -1.64, R) >droYak2.chr2L 16271993 119 - 22324452 GUCUUUGCUAACGUUGGAGUGUGUACUCGCUAACUGGGCAGUGUGUACACACCGCAUACAAUGUAAAAUCACUUACAUUUUCCAGAACUCCAAA-ACUGUACAGUUUGAAUUUUUUAGAG .......((((...(((((((((....)))...((((((.((((....)))).))....(((((((......)))))))..)))).))))))((-(((....))))).......)))).. ( -29.10, z-score = -1.23, R) >droSec1.super_3 2392188 103 + 7220098 GUCUUCGCUAACUUUGGAGUGUGUACUCGCUAACAUGAAAGUGUGUACACACU----------------CGCUAUCAUUUUCCAGAACCCAAAAUCUUGUACAGUUUGAACUUUUAGAG- ...........(((((((((((((((.((((........)))).)))))))))----------------)............((((...(((....))).....))))......)))))- ( -23.50, z-score = -1.43, R) >droSim1.chr2L 6651532 103 + 22036055 GUCUUCGCUAGCUUUGGAGUGUGUACUCGCUAAAAUGGAAGUGUGUACACACU----------------CGCUAUCAUUUUCCUGAACCUAAAAUGUUGUACAGUUUGAAAUUUUAGAG- ......(.((((....((((((((((.((((........)))).)))))))))----------------))))).)............(((((((.(((.......))).)))))))..- ( -27.20, z-score = -2.30, R) >consensus GUCUUCGCUAGCUUUGGAGUGUGUACUCGCUAACAUGGAAGUGUGUACACACUGC_UA_AAUG_AAAA_CGCUAUCAUUUUCCAGAACCCAAAAUCUUGUACAGUUUGAACUUUUAGAG_ .................(((((((((.((((........)))).)))))))))................................................................... (-15.22 = -15.82 + 0.60)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:22:18 2011