| Sequence ID | dm3.chr3R |
|---|---|
| Location | 5,936,821 – 5,936,921 |
| Length | 100 |
| Max. P | 0.998456 |

| Location | 5,936,821 – 5,936,921 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 64.32 |
| Shannon entropy | 0.72433 |
| G+C content | 0.39187 |
| Mean single sequence MFE | -23.26 |
| Consensus MFE | -10.38 |
| Energy contribution | -10.29 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.62 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.36 |
| SVM RNA-class probability | 0.998456 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
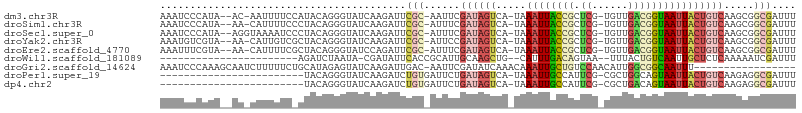
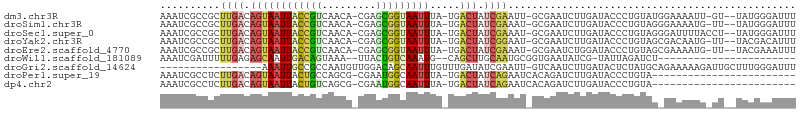
>dm3.chr3R 5936821 100 + 27905053 AAAUCCCAUA--AC-AAUUUUCCAUACAGGGUAUCAAGAUUCGC-AAUUCGAUAGUCA-UAAAUUACCGCUCG-UGUUGACGGUAAUUACUGUCAAGCGGCGAUUU ....(((...--..-.............))).........((((-..((.((((((..-..((((((((.((.-....)))))))))))))))).))..))))... ( -21.77, z-score = -1.60, R) >droSim1.chr3R 12078016 100 - 27517382 AAAUCCCAUA--AA-CAUUUUCCCUACAGGGUAUCAAGAUUCGC-AUUUCGAUAGUCA-UAAAUUACCGCUCG-UGUUGACGGUAAUUACUGUCAAGCGGCGAUUU ..........--..-......(((....))).........((((-.....((((((..-..((((((((.((.-....)))))))))))))))).....))))... ( -23.40, z-score = -2.02, R) >droSec1.super_0 5143883 101 - 21120651 AAAUCCCAUA--AGGUAAAAUCCCUACAGGGUAUCAAGAUUCGC-AUUUCGAUAGUCA-UAAAUUACCGCUCG-UGUUGACGGUAAUUACUGUCAAGCGGCGAUUU (((((.....--.(((((...(((....)))((((.((((....-)))).))))....-....)))))((.((-(.((((((((....)))))))))))))))))) ( -26.80, z-score = -2.59, R) >droYak2.chr3R 9984840 100 + 28832112 AAAUGUCGUA--AA-CAUUGUCGCUACAGGGUAUCAAGAUUCGC-AUUCCGAUAGUCA-UAAAUUACCGCUCG-UGUUGACGGUAAUUACUGUCAAGCGGCGAUUU ..........--..-.(((((((((...(((.............-..)))((((((..-..((((((((.((.-....)))))))))))))))).))))))))).. ( -27.26, z-score = -1.88, R) >droEre2.scaffold_4770 2083356 100 - 17746568 AAAUUUCGUA--AA-CAUUUUCGCUACAGGGUAUCCAGAUUCGC-AUUUCGAUAGUCA-UAAAUUACCGCUCG-UGUUGACGGUAAUUACUGUCAAGCGGCGAUUU ..........--..-.....((((..(.((....))........-.....((((((..-..((((((((.((.-....))))))))))))))))..)..))))... ( -23.10, z-score = -1.27, R) >droWil1.scaffold_181089 1217023 78 - 12369635 -----------------------AGAUCUAAUA-CGAUAUUCACCGCAUUGCAAGCUG--CAUUUGACAGUAA--UUUACUGUCAAUUGCUCUCAAAAAUCGAUUU -----------------------..........-((((.......((.......)).(--((.(((((((((.--..))))))))).)))........)))).... ( -18.50, z-score = -3.05, R) >droGri2.scaffold_14624 2280000 88 + 4233967 AAAUCCCAAAGCAAUCUUUUUCUGCAUAGAGUAUCAAGAUUGAC-AAUUCGAUAUCAAACAAAUUGCUGUCCAACAUUGGCGGCAAUUU----------------- ..(((......(((((((.((((....))))....)))))))..-.....))).......(((((((((.(((....))))))))))))----------------- ( -19.32, z-score = -2.29, R) >droPer1.super_19 1155258 80 - 1869541 ------------------------UACAGGGUAUCAAGAUCUGUGAUUCUGAUAGUCA-UAAAUUGCCAUUCG-CGCUGGCAGUAAUUACUGUCAAGAGGCGAUUU ------------------------....((.(((((..(((...)))..))))).)).-.((((((((..((.-...(((((((....))))))).)))))))))) ( -24.30, z-score = -2.57, R) >dp4.chr2 25785303 80 - 30794189 ------------------------UACAGGGUAUCAAGAUCUGUGAUUCUGAUAGUCA-UAAAUUGCCAUUCG-CGCUGACAGUAAUUACUGUCAAGAGGCGAUUU ------------------------....((.(((((..(((...)))..))))).)).-.((((((((..((.-...(((((((....))))))).)))))))))) ( -24.90, z-score = -3.04, R) >consensus AAAUCCCAUA__AA_CAUUUUC_CUACAGGGUAUCAAGAUUCGC_AUUUCGAUAGUCA_UAAAUUACCGCUCG_UGUUGACGGUAAUUACUGUCAAGCGGCGAUUU ............................((((......))))........((((((.....((((((((.(((....)))))))))))))))))............ (-10.38 = -10.29 + -0.09)
| Location | 5,936,821 – 5,936,921 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 64.32 |
| Shannon entropy | 0.72433 |
| G+C content | 0.39187 |
| Mean single sequence MFE | -20.97 |
| Consensus MFE | -7.42 |
| Energy contribution | -7.31 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.62 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.728858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
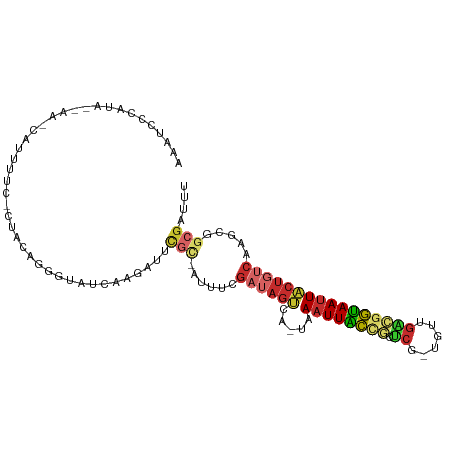
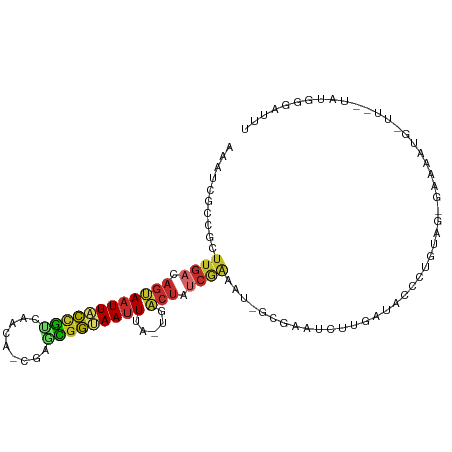
>dm3.chr3R 5936821 100 - 27905053 AAAUCGCCGCUUGACAGUAAUUACCGUCAACA-CGAGCGGUAAUUUA-UGACUAUCGAAUU-GCGAAUCUUGAUACCCUGUAUGGAAAAUU-GU--UAUGGGAUUU ...((((...((((.((((((((((((.....-...)))))))))..-..))).))))...-))))..........((((((.........-..--)))))).... ( -22.10, z-score = -0.88, R) >droSim1.chr3R 12078016 100 + 27517382 AAAUCGCCGCUUGACAGUAAUUACCGUCAACA-CGAGCGGUAAUUUA-UGACUAUCGAAAU-GCGAAUCUUGAUACCCUGUAGGGAAAAUG-UU--UAUGGGAUUU (((((.(((...((((..(((((((((.....-...)))))))))..-....((((((.((-....)).))))))(((....)))....))-))--..)))))))) ( -23.30, z-score = -1.02, R) >droSec1.super_0 5143883 101 + 21120651 AAAUCGCCGCUUGACAGUAAUUACCGUCAACA-CGAGCGGUAAUUUA-UGACUAUCGAAAU-GCGAAUCUUGAUACCCUGUAGGGAUUUUACCU--UAUGGGAUUU ...((((...((((.((((((((((((.....-...)))))))))..-..))).))))...-))))..........(((((((((......)))--)))))).... ( -29.30, z-score = -3.13, R) >droYak2.chr3R 9984840 100 - 28832112 AAAUCGCCGCUUGACAGUAAUUACCGUCAACA-CGAGCGGUAAUUUA-UGACUAUCGGAAU-GCGAAUCUUGAUACCCUGUAGCGACAAUG-UU--UACGACAUUU ...((((..(....((..(((((((((.....-...)))))))))..-))..(((((..((-....))..)))))....)..)))).((((-((--...)))))). ( -21.00, z-score = -0.55, R) >droEre2.scaffold_4770 2083356 100 + 17746568 AAAUCGCCGCUUGACAGUAAUUACCGUCAACA-CGAGCGGUAAUUUA-UGACUAUCGAAAU-GCGAAUCUGGAUACCCUGUAGCGAAAAUG-UU--UACGAAAUUU ...(((((((((((.((((((((((((.....-...)))))))))..-..))).))))...-))).....((....))....)))).....-..--.......... ( -21.40, z-score = -0.96, R) >droWil1.scaffold_181089 1217023 78 + 12369635 AAAUCGAUUUUUGAGAGCAAUUGACAGUAAA--UUACUGUCAAAUG--CAGCUUGCAAUGCGGUGAAUAUCG-UAUUAGAUCU----------------------- .....(((((......(((.(((((((((..--.))))))))).))--).......((((((((....))))-))))))))).----------------------- ( -22.80, z-score = -3.10, R) >droGri2.scaffold_14624 2280000 88 - 4233967 -----------------AAAUUGCCGCCAAUGUUGGACAGCAAUUUGUUUGAUAUCGAAUU-GUCAAUCUUGAUACUCUAUGCAGAAAAAGAUUGCUUUGGGAUUU -----------------(((((((.((((....))).).)))))))..((((((......)-)))))........(((...((((.......))))...))).... ( -17.20, z-score = -0.20, R) >droPer1.super_19 1155258 80 + 1869541 AAAUCGCCUCUUGACAGUAAUUACUGCCAGCG-CGAAUGGCAAUUUA-UGACUAUCAGAAUCACAGAUCUUGAUACCCUGUA------------------------ .............((((.......(((((...-....))))).....-....(((((..(((...)))..)))))..)))).------------------------ ( -13.70, z-score = -0.36, R) >dp4.chr2 25785303 80 + 30794189 AAAUCGCCUCUUGACAGUAAUUACUGUCAGCG-CGAAUGGCAAUUUA-UGACUAUCAGAAUCACAGAUCUUGAUACCCUGUA------------------------ ...((((...((((((((....)))))))).)-)))...........-....(((((..(((...)))..))))).......------------------------ ( -17.90, z-score = -1.71, R) >consensus AAAUCGCCGCUUGACAGUAAUUACCGUCAACA_CGAGCGGUAAUUUA_UGACUAUCGAAAU_GCGAAUCUUGAUACCCUGUAG_GAAAAUG_UU__UAUGGGAUUU ..........((((.((((((((((((.........))))))))).....))).))))................................................ ( -7.42 = -7.31 + -0.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:02:16 2011