
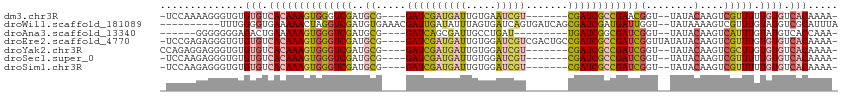
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 5,925,235 – 5,925,333 |
| Length | 98 |
| Max. P | 0.760492 |

| Location | 5,925,235 – 5,925,333 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 75.62 |
| Shannon entropy | 0.46462 |
| G+C content | 0.47129 |
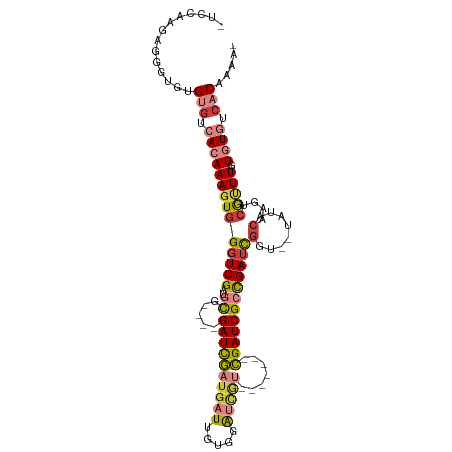
| Mean single sequence MFE | -31.34 |
| Consensus MFE | -16.60 |
| Energy contribution | -17.71 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.760492 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
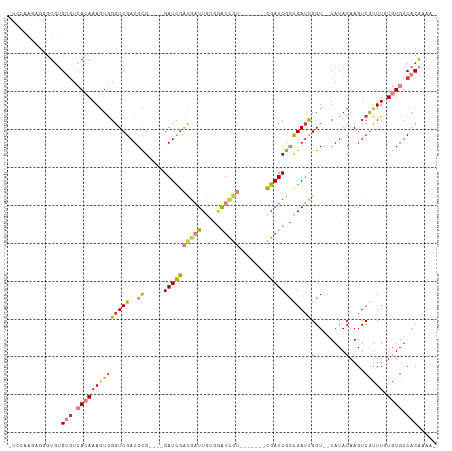
>dm3.chr3R 5925235 98 - 27905053 -UCCAAAAGGGUGUGUGUCACAAAGUGGGUCGAUGCG----GAUCGAUGAUUGUGAAUCGU-------CGAUCGCCGAACGGU--UAUACAAGUCGUUUUUGUGUCACAAAA- -(((....)))..((((.(((((((.....((((..(----((((((((((.....)))))-------)))))(((....)))--....)..)))).))))))).))))...- ( -31.50, z-score = -1.95, R) >droWil1.scaffold_181089 1200831 101 + 12369635 ----------UUUGGGGUGAAAAACUAGGUCGAUGUGAAACGAUUGAUAUUUAGUGAUCAGUGAUCAGCGAUCGAUGAUUGGU--UAUAAAAGUCGUUUGUAUGUCGCAUUUA ----------.....(((.....))).....(((((((.((((.((((.((((.((((((((.(((.......))).))))))--)))))).)))).))))...))))))).. ( -24.30, z-score = -1.34, R) >droAna3.scaffold_13340 6488516 91 - 23697760 ------GGGGGGGAGACUGAAAAAGUGGGUCGAUGCG----GAUCAGCGAUUGCCUGAU---------UGAUCGGCGAUCGGU--UAUACAAGUCAUUUGUAUGUCACCAAA- ------.....((.(((.....(((((((((......----))))..((((((((.((.---------...))))))))))..--.........)))))....))).))...- ( -24.90, z-score = -0.40, R) >droEre2.scaffold_4770 2071698 107 + 17746568 -UCCGAGAGGGUGUGUGUCACAAAGUGGGUCGAUGCG----GAUCGAUGAUUGUGGAUCGUCGACUGCCGAUCGCCGAUCGGUUAUAUACAAGUCGUUUGUGUGUCACAAAA- -(((....)))((((...((((((...(((((..(((----(.(((((((((...))))))))))))))))))..((((..((.....))..))))))))))...))))...- ( -38.30, z-score = -1.76, R) >droYak2.chr3R 9972952 99 - 28832112 CCAGAGGAGGGUGUGUGUCACAAAGUGGGUCGAUGCG----GAUCGAUGAUUGUGGAUCGU-------CGAUCGCCGAUCGGU--UAUACAAGUCGCUUGUGUGUCACAAAA- ((......))...((((.((((((((((((((..(((----.((((((((((...))))))-------))))))))))))(..--....)....))))).)))).))))...- ( -35.20, z-score = -2.05, R) >droSec1.super_0 5132252 98 + 21120651 -UCCAAGAGGGUGUGUGUCACAAAGUGGGUCGAUGCG----GAUCGAUGAUUGUGGAUCGU-------CGAUCGCCGAUCGGU--UAUACAAGUCGUUUUUGUGUCACAAAA- -(((....)))..((((.(((((((..(((((..(((----.((((((((((...))))))-------))))))))))))(..--....).......))))))).))))...- ( -32.60, z-score = -1.87, R) >droSim1.chr3R 12066465 98 + 27517382 -UCCAAGAGGGUGUGUGUCACAAAGUGGGUCGAUGCG----GAUCGAUGAUUGUGGAUCGU-------CGAUCGCCGAUCGGU--UAUACAAGUCGUUUUUGUGUCACAAAA- -(((....)))..((((.(((((((..(((((..(((----.((((((((((...))))))-------))))))))))))(..--....).......))))))).))))...- ( -32.60, z-score = -1.87, R) >consensus _UCCAAGAGGGUGUGUGUCACAAAGUGGGUCGAUGCG____GAUCGAUGAUUGUGGAUCGU_______CGAUCGCCGAUCGGU__UAUACAAGUCGUUUGUGUGUCACAAAA_ ..............(((.((((((((((((((((........))))))(((((..(((((........)))))..))))).............)))))).)))).)))..... (-16.60 = -17.71 + 1.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:02:14 2011