| Sequence ID | dm3.chr3R |
|---|---|
| Location | 5,869,743 – 5,869,845 |
| Length | 102 |
| Max. P | 0.810485 |

| Location | 5,869,743 – 5,869,845 |
|---|---|
| Length | 102 |
| Sequences | 13 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 67.71 |
| Shannon entropy | 0.74989 |
| G+C content | 0.41059 |
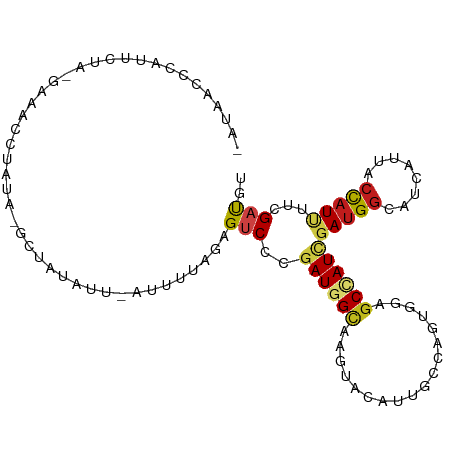
| Mean single sequence MFE | -23.71 |
| Consensus MFE | -8.50 |
| Energy contribution | -8.75 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.810485 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 5869743 102 + 27905053 AAUAACCCAUUUUGAGAAUCCUAUA-GCUGUAUU-AUUCUAGAGUCCCGAUGGCAAGUACAUUGCCAGUGGAGCCAUCGAUGGUAUCAUUACCAUUUUCGAUGU ..............(((((..(((.-...)))..-)))))...(..(((.((((((.....)))))).)))..)(((((((((((....)))))...)))))). ( -27.30, z-score = -2.61, R) >droSim1.chr3R 12011304 102 - 27517382 AAUAACCCAUUCUAAGAAACCUAUA-GCUAUAUU-AUUGUAGAGUCCCGAUGGCAAGUACAUUGCCAGUGGAGCCAUCGAUGGUAUCAUUACCAUUUUCGAUGU .........................-.(((((..-..))))).(..(((.((((((.....)))))).)))..)(((((((((((....)))))...)))))). ( -27.40, z-score = -3.04, R) >droSec1.super_0 5075743 102 - 21120651 AAUAACCCAUUCUAAGAAACCUAUA-GCUAUAUU-AUUCUAGAGUCCCGAUGGCAAGUACAUUGCCAGUGGAGCCAUCGAUGGUAUCAUUACCAUUUUCGAUGU ..........((((.(((....(((-....))).-.)))))))(..(((.((((((.....)))))).)))..)(((((((((((....)))))...)))))). ( -26.90, z-score = -3.02, R) >droYak2.chr3R 9915641 102 + 28832112 AAUAACCCAUCUGCCGAGACCUAUA-GCUAUAUU-AUUUUAGAGUCCUGAUGGCAAGUACAUUGCCAGUGGAGCCAUCGAUGGUAUCAUCACCAUUUUCGAUGU .........((..(((.((((((..-........-....))).))).)).((((((.....)))))))..))..((((((((((......))))...)))))). ( -24.74, z-score = -1.36, R) >droEre2.scaffold_4770 2014507 101 - 17746568 -AAAACCUAUCUGCCAAGACUUUUA-GCUAUACU-AUUUUAGAGUCCUGAUGGCAAGUACAUUGCCAGUGGAGCCAUCGAUGGUAUCAUCACCAUUUUCGAUGU -........((..(((.((((((((-(.....))-)....)))))).)).((((((.....)))))))..))..((((((((((......))))...)))))). ( -26.60, z-score = -2.28, R) >droAna3.scaffold_13340 6430338 100 + 23697760 ---ACUACCCUCCACUCUGUCUAAAUAUC-UUGUAUUUCCAGAGCCCCGAUGGCAAGUACAUAGCCAGUGGGGCUAUAGACGGCAUCAUUACUAUCUUUGACGU ---.............(((((((......-(((......)))(((((((.((((.........)))).))))))).)))))))..................... ( -26.30, z-score = -1.49, R) >dp4.chr2 25711138 103 - 30794189 CGUAGACUGCUCCAUUACACUUAUUCCAU-UUUGCAUUGCAGAGUCCAGAUGGCAAAUACAUAGCCAGCGGAGCCAUUGAUGGCAUCAUCACCAUCUUUGAUGU ........(((((..............((-(((((...)))))))...(.((((.........)))).))))))((..(((((........)))))..)).... ( -25.50, z-score = -0.97, R) >droPer1.super_19 1077655 103 - 1869541 CGUAGACUGCUCCAUUGCAAUUAUUCCAU-UUUGCAUUGCAGAGUCCAGAUGGCAAAUACAUAGCCAGCGGAGCCAUUGAUGGCAUCAUCACCAUCUUUGAUGU ........(((((.(((((((((......-..)).)))))))......(.((((.........)))).))))))((..(((((........)))))..)).... ( -26.70, z-score = -0.70, R) >droWil1.scaffold_181108 3047214 85 - 4707319 -------------------UCUAACCGAGCCAUUACAUUUAGAGUCCUGAUGGUAAAUACGUGGCCAGCGGAGCUAUAGAUGGCAUAAUUACCAUAUUCGAUGU -------------------.........((((((.....(((..(((...((((.........))))..))).)))..)))))).................... ( -16.40, z-score = 0.39, R) >droVir3.scaffold_12855 7380681 99 - 10161210 UGGAAACUACUAUA----AAGUGUACUUU-UUACAUUUUUAGAGUCCUGAUGGCAAGUACAUCGCAAAUGGUGCAAUUGAUGGCAUCAUUACCAUCUUCGAUGU .(((..((.(((.(----(((((((....-.)))))))))))))))).(((((...((.....)).((((((((........)))))))).)))))........ ( -23.10, z-score = -1.35, R) >droMoj3.scaffold_6540 22193139 88 - 34148556 ----------CAGA----AAUUAAA-UUA-UUCCAAAUGUAGAGUCCUGAUGGUAAAUACAUUGCAAACGGUGCAAUCGAUGGCAUCAUUACCAUAUUCGAUGU ----------..((----(......-...-.....((((((....((....))....))))))......(((((........))))).........)))..... ( -14.00, z-score = 0.46, R) >droGri2.scaffold_15074 4147752 90 + 7742996 --------------UAUAGCCUAAAAACCAUUGAAUUUACAGAGUCCGGAUGGAAAGUAUAUAGCGAAUGGUGCCAUCGAUGGCAUCAUUACCAUCUUUGAUGU --------------.............................(((.((((((...((.....)).((((((((((....)))))))))).))))))..))).. ( -21.80, z-score = -1.52, R) >anoGam1.chr2L 47817433 104 + 48795086 AAGGGACGAUAAAAGGAAAGUUUCAUGUUAUACCCAUUUCAGAGCCCCGAUGGCAAGUACAUUGCGAGCGGUGCGAUCGAUGGCAUUAUCAACAUUUUCGAUGU ..(..((............))..)((((((((.(((((.....((.(((.(.((((.....)))).).))).))....)))))...))).)))))......... ( -21.50, z-score = 0.71, R) >consensus _AUAACCCAUUCUA_GAAACCUAUA_GCUAUAUU_AUUUUAGAGUCCCGAUGGCAAGUACAUUGCCAGUGGAGCCAUCGAUGGCAUCAUUACCAUUUUCGAUGU ...........................................(((..((((((..................))))))(((((........)))))...))).. ( -8.50 = -8.75 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:02:08 2011