
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 5,860,606 – 5,860,680 |
| Length | 74 |
| Max. P | 0.608860 |

| Location | 5,860,606 – 5,860,680 |
|---|---|
| Length | 74 |
| Sequences | 10 |
| Columns | 84 |
| Reading direction | forward |
| Mean pairwise identity | 66.75 |
| Shannon entropy | 0.68448 |
| G+C content | 0.38923 |
| Mean single sequence MFE | -14.47 |
| Consensus MFE | -3.63 |
| Energy contribution | -4.26 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.25 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.608860 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 5860606 74 + 27905053 UUGCAUUG--------CCAAUAUCCGCUGGCAAUGCAAUCAAGCAGUAAAUA-AAUUCGAAAUAGUAACA-AUACAGAAACACC ((((((((--------(((........)))))))))))..............-...........(((...-.)))......... ( -16.50, z-score = -3.27, R) >droSim1.chr3R 12001866 74 - 27517382 UUGCAUUG--------CCAAUAUCCGCUGGCAAUGCAAUCAAGCAGUAAAUA-AAUUCGAAAUAGUAACA-AUACAGAAACACC ((((((((--------(((........)))))))))))..............-...........(((...-.)))......... ( -16.50, z-score = -3.27, R) >droSec1.super_0 5066276 74 - 21120651 UUGCAUUG--------CCAAUAUCCGCUGGCAAUGCAAUCAAGCAGUAAAUA-AAUUCGAAAUAGUAACA-AUACAGAAACACC ((((((((--------(((........)))))))))))..............-...........(((...-.)))......... ( -16.50, z-score = -3.27, R) >droYak2.chr3R 9905987 66 + 28832112 UUGCAUUG--------CC-------GCUGGCAAUGCAAUCAAGCAGUAAAUA-AAUUCGAAA-AGUUACA-AUACAGAAACACC ((((((((--------((-------...))))))))))..............-.........-.......-............. ( -13.70, z-score = -2.05, R) >droEre2.scaffold_4770 2004659 74 - 17746568 UUGCAUUG--------CCAAUAUCCGCUGGCAAUGCAAUCAAGCAGUAAAUA-AAUUCGAAAUAGUAACA-AUACAAAAACAUC ((((((((--------(((........)))))))))))..............-...........(((...-.)))......... ( -16.50, z-score = -3.36, R) >droAna3.scaffold_13340 6420774 64 + 23697760 UUGCAUUG--------CCAAUAUCCGCUGGCAAUGCAAUCAAGCAGCAAACA-AAAACAGCUCCACCGAA-ACC---------- ((((((((--------(((........)))))))))))....(.(((.....-......))).)......-...---------- ( -17.60, z-score = -3.63, R) >droWil1.scaffold_181089 5511637 73 + 12369635 UUGCAUUGG-------CCAAUAUCCGCUGGCAAUGCAUUCAAGCAGCAACAA-AAAGC-AAACAACAACA-ACAAAUAAAAAU- .(((((((.-------(((........))))))))))........((.....-...))-...........-............- ( -13.10, z-score = -1.16, R) >droGri2.scaffold_15074 4128513 79 + 7742996 UGCUACUAC--UGCCGCCAAUAUCCGCUGACGAUGCAUUCAAGCGGUAACAACAACAAAAACUUACCAUUAAGGGCACAAC--- .((......--.)).(((.....(((((...((.....)).)))))...............((((....))))))).....--- ( -14.30, z-score = -0.98, R) >droVir3.scaffold_13047 13099647 78 + 19223366 UUGCAUUCGACUACUGCCAAUAUCCGCUGUGAGUGCAGUCAAGCGGCAA-CA-AAAAGAAGACAACAACA-ACAAAACGGA--- .(((((((.((....((........)).)))))))))(((....)))..-..-.................-..........--- ( -11.60, z-score = 0.94, R) >droMoj3.scaffold_6540 31149938 71 + 34148556 ----ACUG--------UCAAUAUCCGUUGGCAAUGCAUUCAAGCGGCAAUAA-AAAAAAAACUAGAUGUGUAUAAUAUGGAAAC ----..((--------(((((....)))))))((((((((.((.........-........)).)).))))))........... ( -8.43, z-score = 0.46, R) >consensus UUGCAUUG________CCAAUAUCCGCUGGCAAUGCAAUCAAGCAGUAAAUA_AAUUCGAAAUAGUAACA_AUACAAAAACACC .(((((((........(((........))))))))))............................................... ( -3.63 = -4.26 + 0.63)
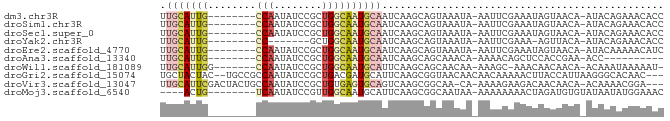

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:02:07 2011