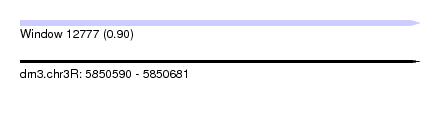
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 5,850,590 – 5,850,681 |
| Length | 91 |
| Max. P | 0.897363 |

| Location | 5,850,590 – 5,850,681 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 73.21 |
| Shannon entropy | 0.51856 |
| G+C content | 0.30676 |
| Mean single sequence MFE | -11.79 |
| Consensus MFE | -9.12 |
| Energy contribution | -9.06 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.17 |
| Mean z-score | -0.89 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.897363 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 5850590 91 + 27905053 AACUUGGCCAAUGGCCAGGAAAUUUAUAACAAAU-AUUCGAAACAC-CAACAUCAUCUCGUAGUCCCAUUCAUAAACAAAAUAAUUAAUUUUG ..(((((((...)))))))...............-...........-.............................((((((.....)))))) ( -12.00, z-score = -1.47, R) >droSim1.chr3R 11992013 89 - 27517382 AACUUGGCCAAUGGCCAGGAAAUUUAUAACAAGUUAUUCGAAACAC-CAACAUC---UCGUAGUCCCAUUCAUAAACAAAAUAAUUAAUUUUG ..(((((((...))))))).............(..((((((.....-.......---))).)))..).........((((((.....)))))) ( -12.50, z-score = -1.25, R) >droSec1.super_0 5052852 89 - 21120651 AACUUGGCCAAUGGCCAGGAAAUUUAUAACAAGUUAUUCGAAACAC-CAACAUC---UCGUAGUCCCAUUCAUAAACAAAAUAAUUAAUUUUG ..(((((((...))))))).............(..((((((.....-.......---))).)))..).........((((((.....)))))) ( -12.50, z-score = -1.25, R) >droYak2.chr3R 9895807 88 + 28832112 AACUUGGCCAAUGGCCAGGAAAUUUAUAAUAGAU-AUUGGAAGCAA-CAACAUC---UUGUAGUCCCAUUCAUAAACAGAAUAAUUAAUUUUC ..(((((((...)))))))............((.-..(((..((.(-(((....---)))).)).))).))...................... ( -15.80, z-score = -1.22, R) >droEre2.scaffold_4770 1994564 88 - 17746568 AACUUGGCCAAUGGCCAGGAAAUUUAUAUUAAAU-AUUCGAAGCAG-CAACAUC---UUGUAGUCCCAUUCACAAACAAAAUAAUUAAUUUUG ..(((((((...)))))))...............-....(((((.(-(((....---)))).))....))).....((((((.....)))))) ( -14.20, z-score = -1.27, R) >droAna3.scaffold_13340 6411177 89 + 23697760 AACUUGGCCAAUAGCCAGGAAAUUCAUACGAAAU-AUUCGAAACACACCAAAUC---UUGUUGCCUCAUUCAUAAACAAAAUAAUUAAUUUUG ..((((((.....)))))).........(((...-..)))..............---...................((((((.....)))))) ( -10.90, z-score = -1.10, R) >droVir3.scaffold_13047 13086838 76 + 19223366 GAAUUGGCCAAUAGCCAGUAAAUU--UCAUAAUU-AUUAAAAA-UA-CAAUAUU--UGU-GCGCAACAUUAAUUUUUCGGAUUG--------- ..((((((.....)))))).....--.(((((.(-(((.....-..-.)))).)--)))-).......................--------- ( -9.40, z-score = 0.09, R) >droMoj3.scaffold_6540 31136254 78 + 34148556 GAAUUGGCUAAUUGCCAGCAAAUU--UCAUAAUU-AUUAAAAAAUA-CAACAUU--UUUCGCGCAACAUUAAUUUUUCAAAUUG--------- (((.((((.....))))((.((((--....))))-....((((((.-....)))--))).)).............)))......--------- ( -7.00, z-score = 0.38, R) >consensus AACUUGGCCAAUGGCCAGGAAAUUUAUAAUAAAU_AUUCGAAACAC_CAACAUC___UUGUAGUCCCAUUCAUAAACAAAAUAAUUAAUUUUG ..((((((.....)))))).......................................................................... ( -9.12 = -9.06 + -0.06)

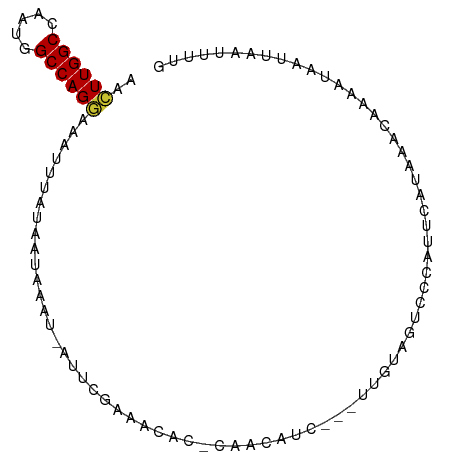

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:02:06 2011