
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 5,840,998 – 5,841,118 |
| Length | 120 |
| Max. P | 0.806006 |

| Location | 5,840,998 – 5,841,118 |
|---|---|
| Length | 120 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.47 |
| Shannon entropy | 0.40882 |
| G+C content | 0.54754 |
| Mean single sequence MFE | -41.28 |
| Consensus MFE | -27.47 |
| Energy contribution | -27.52 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.806006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
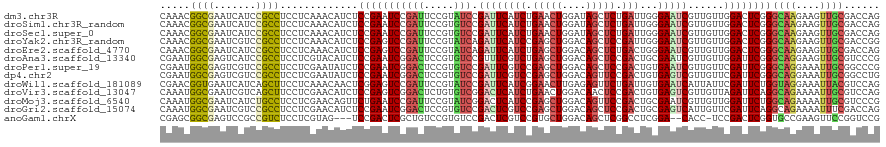
>dm3.chr3R 5840998 120 + 27905053 CAAACGGCGAAUCAUCCGCCUCCUCAAACAUCUCCGAAUCCGAUUCCGUAUCCGAUUCAUCUGAACUGGAUAGCUCUGAUUGGGAAUCGUUGUUGGACUCGGGCAAGAAGUUGCGACCAG .....((((.......)))).............((((.((((((..((((((((.(((....))).)))))).(((.....)))...))..)))))).))))((((....))))...... ( -34.10, z-score = -0.58, R) >droSim1.chr3R_random 387083 120 + 1307089 CAAACGGCGAAUCAUCCGCCUCCUCAAACAUCUCCGAAUCCGAUUCCGUGUCCGAUUCAUCUGAACUGGAUAGCUCUGAUUGGGAAUCGUUGUUGGACUCGGGCAAGAAGUUGCGACCAG .....((((.......)))).............((((.((((((..((((((((.(((....))).)))))).(((.....)))...))..)))))).))))((((....))))...... ( -34.10, z-score = -0.26, R) >droSec1.super_0 5043487 120 - 21120651 CAAACGGCGAAUCAUCCGCCUCCUCAAACAUCUCCGAAUCCGAUUCCGUGUCCGAUUCAUCUGAACUGGAUAGCUCUGAUUGGGAAUCGUUGUUGGACUCGGGCAAGAAGUUGCGACCAG .....((((.......)))).............((((.((((((..((((((((.(((....))).)))))).(((.....)))...))..)))))).))))((((....))))...... ( -34.10, z-score = -0.26, R) >droYak2.chr3R_random 1252785 120 + 3277457 CAAACGGCGAAUCGUCCGCCUCCUCAAACAUCUCCGAGUCCGAUUCCGUAUCAGAUUCAUCCGAGCUGGACAGCUCCGAUUGGGAAUCGUUGUUGGACUCGGGCAAGAAGUUGCGACCGG .....((((.......)))).((..........(((((((((((..((..((......(((.(((((....))))).)))...))..))..)))))))))))((((....))))....)) ( -40.70, z-score = -0.91, R) >droEre2.scaffold_4770 1984981 120 - 17746568 CAAACGGCGAAUCAUCCGCCUCCUCAAACAUCUCCGAGUCCGAUUCCGUAUCAGAUUCAUCUGAGCUGGACAGCUCUGACUGGGAAUCGUUGUUGGACUCGGGCAAGAAGUUGCGACCAG .....((((.......)))).............(((((((((((..((..((((..(((...(((((....))))))))))))....))..)))))))))))((((....))))...... ( -41.80, z-score = -2.00, R) >droAna3.scaffold_13340 6401868 120 + 23697760 CGAAUGGCGAGUCAUCCGCCUCCUCGUACAUCUCCGAAUCGGACUCCGUGUCCGUUUCGUCUGAGCUGGACAGCUCCGACUGCGAAUCGUUGUUGGAUUCGGGCAGGAAGUUGCGUCCCG .....((((.......))))....((((.(..((((((.(((((.....))))).)))(((((((((....)))((((((.(((...))).)))))).)))))).)))..)))))..... ( -42.60, z-score = -0.54, R) >droPer1.super_19 1038667 120 - 1869541 CGAAUGGCGAGUCGUCCGCCUCCUCGAAUAUCUCCGAAUCGGACUCCGUGUCCGAUUCGUCCGAGCUGGACAGCUCCGACUGUGAAUCGUUGUUCGAUUCGGGCAGGAAAUUGCGGCCCG (((..((((.......))))...))).........(((((((((.....)))))))))(((.(((((....))))).)))...((((((.....))))))((((.(.......).)))). ( -49.00, z-score = -2.10, R) >dp4.chr2 25671544 120 - 30794189 CGAAUGGCGAGUCGUCCGCCUCCUCGAAUAUCUCCGAAUCGGACUCCGUGUCCGAUUCGUCCGAGCUGGACAGUUCCGACUGUGAGUCGUUGUUCGAUUCGGGCAGGAAAUUGCGGCCUG .....(((..((..(((((((..(((((((.(((((((((((((.....)))))))))(((.(((((....))))).))).).)))....)))))))...)))).)))....)).))).. ( -49.10, z-score = -2.22, R) >droWil1.scaffold_181089 6832174 120 - 12369635 CGAACGGUGAAUCAUCAGCUUCCUCAAACAACUCGGAGUCCGAUUCCGUAUCCGAUUCAUCGGAACUUGAGAGUUCUGAUUGUGAAUCAUUAUUCGAUUCUGGUAGGAAAUUACGUCCAG ((((..((((.(((((((..(.(((((......((((((...))))))..(((((....)))))..))))).)..))))...))).))))..))))...((((..(.......)..)))) ( -33.20, z-score = -0.99, R) >droVir3.scaffold_13047 13076357 120 + 19223366 CAAAUGGCGAAUCGUCAGCUUCCUCGAACAUCUCCGAGUCGGACUCUGUGUCGGACUCAUCUGAACUGGACAACUCCGACUGUGAGUCGUUGUUAGAUUCAGGCAGAAAAUUGCGUCCAG ....(((((((((..((((...((((........))))...(((((...((((((....(((.....)))....))))))...)))))))))...)))))..((((....))))).))). ( -37.90, z-score = -1.08, R) >droMoj3.scaffold_6540 31126000 120 + 34148556 CAAAUGGCGAAUCAUCUGCCUCCUCGAACAGUUCUGAAUCCGAUUCCGUAUCGGACUCAUCCGAGCUGGACAGUUCCGACUGCGAAUCGUUGUUGGAUUCUGGCAGAAAAUUGCGUCCCG .....(((((((..((((((.....(..(((((((((.((((((.....)))))).)))...))))))..)((.((((((.(((...))).))))))..))))))))..))).))))... ( -42.80, z-score = -3.08, R) >droGri2.scaffold_15074 4100378 120 + 7742996 CAAAUGGCGAAUCGUCCGCCUCCUCGAACAUCUCCGAAUCGGACUCCGUGUCCGACUCGUCCGAGCUGGACAGCUCCGACUGCGAGUCAUUGUUCGAUUCAGGCAGAAAAUUUCGACCAG ....(((((((...((.((((..(((((((.((((((.((((((.....)))))).))(((.(((((....))))).))).).)))....)))))))...)))).))....)))).))). ( -50.10, z-score = -4.43, R) >anoGam1.chrX 18004448 114 - 22145176 CGAGCGGCGAGUCCGCCGUCUCCUCGUAG---UCCGACUCGCUGUCCGUGUCCGACUCGUCCGUGCUGGACAGCUCGGCCUCGGA--CACC-UCCGACUCGGUGCCGAAGUUCCGGUCCG .((((((((....))))).))).......---.((((.(((.((((((.(.((((((.((((.....)))))).)))).).))))--))..-..))).)))).((((......))))... ( -47.20, z-score = -0.54, R) >consensus CAAACGGCGAAUCAUCCGCCUCCUCGAACAUCUCCGAAUCCGAUUCCGUGUCCGAUUCAUCCGAGCUGGACAGCUCCGACUGGGAAUCGUUGUUGGAUUCGGGCAGGAAGUUGCGACCAG .....((((.......)))).............(((((((((((.....))).((((((((.(((((....))))).)))...)))))......))))))))((((....))))...... (-27.47 = -27.52 + 0.05)

| Location | 5,840,998 – 5,841,118 |
|---|---|
| Length | 120 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.47 |
| Shannon entropy | 0.40882 |
| G+C content | 0.54754 |
| Mean single sequence MFE | -43.05 |
| Consensus MFE | -25.79 |
| Energy contribution | -25.26 |
| Covariance contribution | -0.53 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.630678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
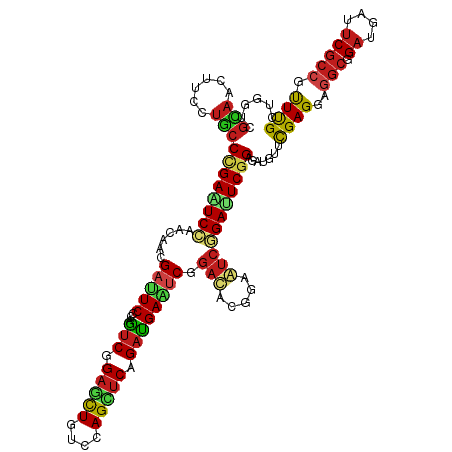
>dm3.chr3R 5840998 120 - 27905053 CUGGUCGCAACUUCUUGCCCGAGUCCAACAACGAUUCCCAAUCAGAGCUAUCCAGUUCAGAUGAAUCGGAUACGGAAUCGGAUUCGGAGAUGUUUGAGGAGGCGGAUGAUUCGCCGUUUG ..((((((..((((..((((((((((.....((((((...(((.(((((....))))).)))))))))(((.....)))))))))))....))..))))..)))((....)))))..... ( -38.60, z-score = -0.89, R) >droSim1.chr3R_random 387083 120 - 1307089 CUGGUCGCAACUUCUUGCCCGAGUCCAACAACGAUUCCCAAUCAGAGCUAUCCAGUUCAGAUGAAUCGGACACGGAAUCGGAUUCGGAGAUGUUUGAGGAGGCGGAUGAUUCGCCGUUUG ..((((((..((((..((((((((((.....((((((...(((.(((((....))))).)))))))))((.......))))))))))....))..))))..)))((....)))))..... ( -36.80, z-score = -0.27, R) >droSec1.super_0 5043487 120 + 21120651 CUGGUCGCAACUUCUUGCCCGAGUCCAACAACGAUUCCCAAUCAGAGCUAUCCAGUUCAGAUGAAUCGGACACGGAAUCGGAUUCGGAGAUGUUUGAGGAGGCGGAUGAUUCGCCGUUUG ..((((((..((((..((((((((((.....((((((...(((.(((((....))))).)))))))))((.......))))))))))....))..))))..)))((....)))))..... ( -36.80, z-score = -0.27, R) >droYak2.chr3R_random 1252785 120 - 3277457 CCGGUCGCAACUUCUUGCCCGAGUCCAACAACGAUUCCCAAUCGGAGCUGUCCAGCUCGGAUGAAUCUGAUACGGAAUCGGACUCGGAGAUGUUUGAGGAGGCGGACGAUUCGCCGUUUG .((..(((..((((..((((((((((......((((((..((((((..(((((.....)))))..))))))..))))))))))))))....))..))))..)))..))............ ( -46.50, z-score = -1.96, R) >droEre2.scaffold_4770 1984981 120 + 17746568 CUGGUCGCAACUUCUUGCCCGAGUCCAACAACGAUUCCCAGUCAGAGCUGUCCAGCUCAGAUGAAUCUGAUACGGAAUCGGACUCGGAGAUGUUUGAGGAGGCGGAUGAUUCGCCGUUUG ..((((((..((((..((((((((((......((((((..(((.(((((....)))))(((....))))))..))))))))))))))....))..))))..)))((....)))))..... ( -42.40, z-score = -1.44, R) >droAna3.scaffold_13340 6401868 120 - 23697760 CGGGACGCAACUUCCUGCCCGAAUCCAACAACGAUUCGCAGUCGGAGCUGUCCAGCUCAGACGAAACGGACACGGAGUCCGAUUCGGAGAUGUACGAGGAGGCGGAUGACUCGCCAUUCG ......(((.((((.....((((((.......))))))..(((.(((((....))))).)))(((.(((((.....))))).))))))).))).((((..(((((.....))))).)))) ( -41.60, z-score = -0.66, R) >droPer1.super_19 1038667 120 + 1869541 CGGGCCGCAAUUUCCUGCCCGAAUCGAACAACGAUUCACAGUCGGAGCUGUCCAGCUCGGACGAAUCGGACACGGAGUCCGAUUCGGAGAUAUUCGAGGAGGCGGACGACUCGCCAUUCG ..(((....((((((.....((((((.....))))))...(((.(((((....))))).)))(((((((((.....)))))))))))))))....(((..(.....)..))))))..... ( -50.80, z-score = -2.77, R) >dp4.chr2 25671544 120 + 30794189 CAGGCCGCAAUUUCCUGCCCGAAUCGAACAACGACUCACAGUCGGAACUGUCCAGCUCGGACGAAUCGGACACGGAGUCCGAUUCGGAGAUAUUCGAGGAGGCGGACGACUCGCCAUUCG ..(((.(((......)))..((.(((.....((((.....))))...(((.((..((((((((((((((((.....))))))))))......))))))..))))).))).)))))..... ( -43.40, z-score = -1.44, R) >droWil1.scaffold_181089 6832174 120 + 12369635 CUGGACGUAAUUUCCUACCAGAAUCGAAUAAUGAUUCACAAUCAGAACUCUCAAGUUCCGAUGAAUCGGAUACGGAAUCGGACUCCGAGUUGUUUGAGGAAGCUGAUGAUUCACCGUUCG ..(((((((......)))..(((((((((....))))...(((((...(((((((.(((((....)))))..((((.......)))).....)))))))...))))))))))...)))). ( -32.70, z-score = -0.84, R) >droVir3.scaffold_13047 13076357 120 - 19223366 CUGGACGCAAUUUUCUGCCUGAAUCUAACAACGACUCACAGUCGGAGUUGUCCAGUUCAGAUGAGUCCGACACAGAGUCCGACUCGGAGAUGUUCGAGGAAGCUGACGAUUCGCCAUUUG .(((.((((......)))..(((((.......(((((...((((((.(..((.......))..).))))))...)))))...((((((....)))))).........))))))))).... ( -37.10, z-score = -0.74, R) >droMoj3.scaffold_6540 31126000 120 - 34148556 CGGGACGCAAUUUUCUGCCAGAAUCCAACAACGAUUCGCAGUCGGAACUGUCCAGCUCGGAUGAGUCCGAUACGGAAUCGGAUUCAGAACUGUUCGAGGAGGCAGAUGAUUCGCCAUUUG ..((.((.((((.((((((.(((((.......)))))(((((....)))))....((((((((((((((((.....))))))))))......))))))..)))))).))))))))..... ( -43.30, z-score = -2.41, R) >droGri2.scaffold_15074 4100378 120 - 7742996 CUGGUCGAAAUUUUCUGCCUGAAUCGAACAAUGACUCGCAGUCGGAGCUGUCCAGCUCGGACGAGUCGGACACGGAGUCCGAUUCGGAGAUGUUCGAGGAGGCGGACGAUUCGCCAUUUG .(((.((((.((.(((((((...(((((((....(((...(((.(((((....))))).)))(((((((((.....))))))))).))).)))))))..))))))).))))))))).... ( -58.40, z-score = -4.67, R) >anoGam1.chrX 18004448 114 + 22145176 CGGACCGGAACUUCGGCACCGAGUCGGA-GGUG--UCCGAGGCCGAGCUGUCCAGCACGGACGAGUCGGACACGGACAGCGAGUCGGA---CUACGAGGAGACGGCGGACUCGCCGCUCG (.(((((...(((((((.....))))))-).((--((((.(.((((..(((((.....)))))..)))).).)))))).)).))).).---.......(((.(((((....)))))))). ( -51.30, z-score = -0.69, R) >consensus CUGGUCGCAACUUCCUGCCCGAAUCCAACAACGAUUCCCAGUCGGAGCUGUCCAGCUCAGAUGAAUCGGACACGGAAUCGGAUUCGGAGAUGUUCGAGGAGGCGGAUGAUUCGCCGUUUG ......(((......)))((((((((......(((((...(((.(((((....))))).)))))))).(((.....))))))))))).......((((..(((.((....))))).)))) (-25.79 = -25.26 + -0.53)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:02:04 2011