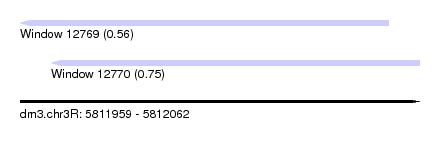
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 5,811,959 – 5,812,062 |
| Length | 103 |
| Max. P | 0.745812 |

| Location | 5,811,959 – 5,812,054 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 71.89 |
| Shannon entropy | 0.50539 |
| G+C content | 0.36565 |
| Mean single sequence MFE | -16.16 |
| Consensus MFE | -11.10 |
| Energy contribution | -11.22 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.17 |
| Mean z-score | -0.61 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.561960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
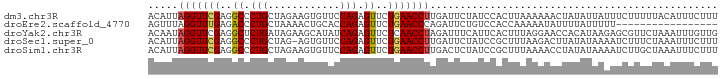
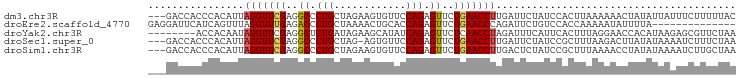
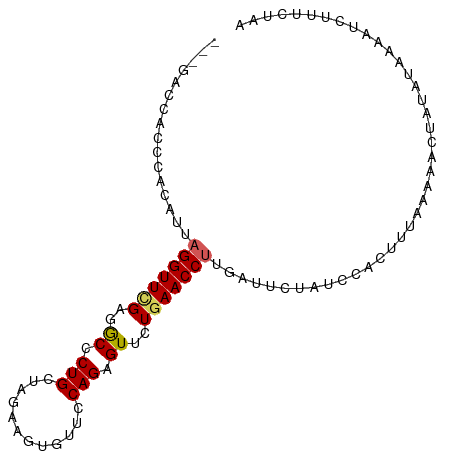
>dm3.chr3R 5811959 95 - 27905053 ACAUUAGGUUCGAGGCCCUGCUAGAAGUGUUCCAGAGUUCUGAACCUUGAUUCUAUCCACUUAAAAAACUAUAUUAUUUCUUUUUACAUUUCUUU .((..(((((((..((.(((............))).))..))))))))).............................................. ( -12.80, z-score = 0.33, R) >droEre2.scaffold_4770 1955872 78 + 17746568 AGUUUAGGUUUGAGACCCUGCUAAAACUGCACCAGAGUUCUGAACCCAGAUUCUGUCCACCAAAAAUAUUUUAUUUUU----------------- ......(((..((.....(((.......))).((((((.(((....))))))))))).))).................----------------- ( -12.60, z-score = -0.80, R) >droYak2.chr3R 9860039 95 - 28832112 ACAAUAGGUUCGAGGCUCUGAUAGAAGCAUAUCAGAGUUCUCAACCUAGAUUUCAUUCACUUUAGGAACCACAUAAGAGCGUUCUAAAUUUGUUG ((((((((((.((((((((((((......)))))))).))))))))))................((((((........).)))))....)))).. ( -26.60, z-score = -3.39, R) >droSec1.super_0 5014526 94 + 21120651 ACAUUAGGUUCGAGGCCCUGCUAG-AGUGUUCCAGAGUUCUGAACCUUGAUUCUAUCCGCUUUAAGACUUAUAUAAAAUCUUUCUAAAUUUCUUU ....((((((.(((((...(.(((-(((((((.((....))))))....)))))).).)))))..))))))........................ ( -13.70, z-score = 0.81, R) >droSim1.chr3R 11963283 95 + 27517382 ACAUUAGGUUCGAGGCCCUGCUAGAAGUGUUCCAGAGUUCUGAACCUUGACUCUAUCCGCUUUAAAACCUAUAUAAAAUCUUGCUAAAUUUCUUU ....((((((.(((((...(.((((((.((((.((....))))))))....)))).).)))))..))))))........................ ( -15.10, z-score = -0.01, R) >consensus ACAUUAGGUUCGAGGCCCUGCUAGAAGUGUUCCAGAGUUCUGAACCUUGAUUCUAUCCACUUUAAAAACUAUAUAAAAUCUUUCUAAAUUUCUUU .....(((((((..((.(((............))).))..)))))))................................................ (-11.10 = -11.22 + 0.12)
| Location | 5,811,967 – 5,812,062 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 70.17 |
| Shannon entropy | 0.53343 |
| G+C content | 0.40814 |
| Mean single sequence MFE | -17.10 |
| Consensus MFE | -11.10 |
| Energy contribution | -11.22 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.17 |
| Mean z-score | -0.81 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.745812 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
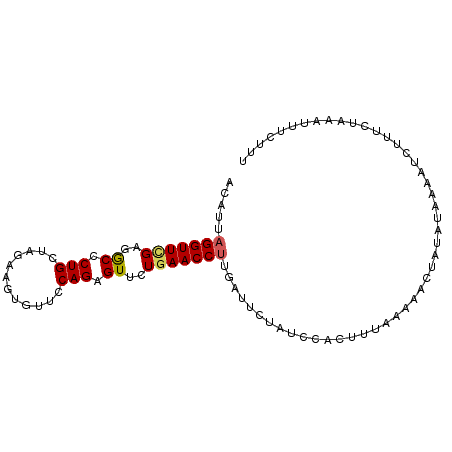
>dm3.chr3R 5811967 95 - 27905053 ---GACCACCCACAUUAGGUUCGAGGCCCUGCUAGAAGUGUUCCAGAGUUCUGAACCUUGAUUCUAUCCACUUAAAAAACUAUAUUAUUUCUUUUUAC ---.........((..(((((((..((.(((............))).))..)))))))))...................................... ( -12.80, z-score = 0.41, R) >droEre2.scaffold_4770 1955877 84 + 17746568 GAGGAUUCAUCAGUUUAGGUUUGAGACCCUGCUAAAACUGCACCAGAGUUCUGAACCCAGAUUCUGUCCACCAAAAAUAUUUUA-------------- ..(((.....((((((.(((..(....)..))).))))))...((((((.(((....))))))))))))...............-------------- ( -17.80, z-score = -1.24, R) >droYak2.chr3R 9860047 90 - 28832112 --------ACCACAAUAGGUUCGAGGCUCUGAUAGAAGCAUAUCAGAGUUCUCAACCUAGAUUUCAUUCACUUUAGGAACCACAUAAGAGCGUUCUAA --------.......((((((.((((((((((((......)))))))).))))))))))................((((((........).))))).. ( -26.10, z-score = -4.01, R) >droSec1.super_0 5014534 94 + 21120651 ---GACCACCCACAUUAGGUUCGAGGCCCUGCUAG-AGUGUUCCAGAGUUCUGAACCUUGAUUCUAUCCGCUUUAAGACUUAUAUAAAAUCUUUCUAA ---............((((((.(((((...(.(((-(((((((.((....))))))....)))))).).)))))..))))))................ ( -13.70, z-score = 0.78, R) >droSim1.chr3R 11963291 95 + 27517382 ---GACCACCCACAUUAGGUUCGAGGCCCUGCUAGAAGUGUUCCAGAGUUCUGAACCUUGACUCUAUCCGCUUUAAAACCUAUAUAAAAUCUUGCUAA ---............((((((.(((((...(.((((((.((((.((....))))))))....)))).).)))))..))))))................ ( -15.10, z-score = 0.01, R) >consensus ___GACCACCCACAUUAGGUUCGAGGCCCUGCUAGAAGUGUUCCAGAGUUCUGAACCUUGAUUCUAUCCACUUUAAAAACUAUAUAAAAUCUUUCUAA ................(((((((..((.(((............))).))..)))))))........................................ (-11.10 = -11.22 + 0.12)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:02:00 2011