
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 5,806,456 – 5,806,561 |
| Length | 105 |
| Max. P | 0.984455 |

| Location | 5,806,456 – 5,806,561 |
|---|---|
| Length | 105 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 69.02 |
| Shannon entropy | 0.65143 |
| G+C content | 0.49109 |
| Mean single sequence MFE | -35.62 |
| Consensus MFE | -13.95 |
| Energy contribution | -13.67 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.984455 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
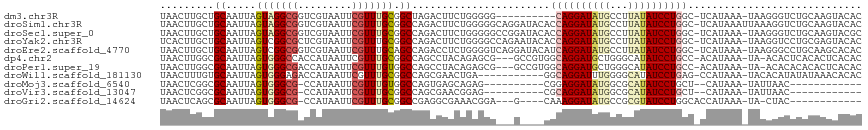
>dm3.chr3R 5806456 105 - 27905053 UAACUUGCUGCAAUUAGUAGGCGGUCGUAAUUCGUUUGCGGCUAGACUUCUGGGGG----------CAGGAUAUGCCUUAUAUCCUGGC-UCAUAAA-UAAGGGUCUGCAAGUACAC ..((((((.((.....((((((((.......)))))))).)).(((((..(((((.----------(((((((((...))))))))).)-)).....-))..))))))))))).... ( -36.60, z-score = -2.87, R) >droSim1.chr3R 11957731 116 + 27517382 UAACUUGCUGCAAUUAGUAGGCGGUCGUAAUUCGUUUGCGGCCAGACUUCUGGGGGCAGGAUACACCAGGAUAUGCCUUAUAUCCUGGC-UCAUAAAUUAAAGGUCUGCAAGUACAC ..(((((((((..((((.((..((((((((.....))))))))...)).))))..))).......((((((((((...)))))))))).-.................)))))).... ( -39.30, z-score = -2.71, R) >droSec1.super_0 5008978 115 + 21120651 UAACUUGCUGCAAUUAGUAGGCGGUCGUAAUUCGUUUGCGGCCAGACUUCUGGGGGCCGGAUACACCAGGAUAUGCCUUAUAUCCUGGC-UCAUAAA-UAAGGGUCUGCAAGUACGC ..((((((..............((((((((.....))))))))(((((..((.(((((((.....))((((((((...)))))))))))-)).....-))..))))))))))).... ( -42.20, z-score = -2.60, R) >droYak2.chr3R 9854540 115 - 28832112 UCACUUGCUGCAAUUAGUCGGCGCUCGUAAUUCGUUUGCGGCCAGACUUCUGGGGCCAGAAUACACCAGGAUAUGCCUUAUAUCCUGGC-UCAUAAA-UAAGGUCCUGCGAGUACAC ..((((((.......(((((((...(((((.....)))))))).))))...((((((.(....).((((((((((...)))))))))).-.......-...)))))))))))).... ( -38.60, z-score = -2.39, R) >droEre2.scaffold_4770 1950398 115 + 17746568 UAACUUGCUGCAAUUAGUCGGCGGUCGUAAUUCGUUUGCAGCCAGACCUCUGGGGUCAGGAUACAUCAGGAUAUGCCUUAUAUCCUGGC-UCAUAAA-UAAGGGCCUGCAAGCACAC ....(((((((.....).)))))).........((((((((((.........(((((((((((....(((.....)))..)))))))))-)).....-....)).)))))))).... ( -38.77, z-score = -2.25, R) >dp4.chr2 25627300 111 + 30794189 UAACUUGGCGCAAUUAGUGGGCCACCAUAAUUCGUUUGCGGCCAGCCUACAGAGCG---GCCGUGGCAGGAUGCUGGGCAUAUCCUGCC-ACAUAAA-UA-ACACUCACACUCACAC .....((((.((.....)).)))).........(((((.((((.(((....).)))---)))(((((((((((.......)))))))))-)).))))-).-................ ( -36.70, z-score = -1.68, R) >droPer1.super_19 992330 111 + 1869541 UAACUUGGCGCAAUUAGUGGGCGACCAUAAUUCGUUUGUGGCCAGCCUACAGAGCG---GCCGUGGCAGGAUGCUGGGCAUAUCCUGCC-ACAUAAA-UA-ACACACACACUCACAC ......(((((..((.((((((..((((((.....))))))...)))))).)))).---)))(((((((((((.......)))))))))-)).....-..-................ ( -41.50, z-score = -3.17, R) >droWil1.scaffold_181130 405075 104 + 16660200 UAACUUUGUGCAAUUAGUGGGAGACCAUAAUUCGUUUGCGGCCAGCGAACUGA-----------GGCAGGAUUUGGGGCAUAUCCUGAG-CCAUAAA-UACACAUAUAUAAACACAC ......((((......((((....)))).....((((((.....))))))...-----------((((((((.((...)).)))))..)-)).....-..))))............. ( -26.10, z-score = -1.39, R) >droMoj3.scaffold_6540 31089497 91 - 34148556 UAACUCGGCGCAAUUAGUGGGCG-CCAUAAUUCGUUUGUGGCCAGUGAGCAGAG----------CGGAGGAUAUGGCGCAUAUCCUGCU--CAUAAA-UAUUAAC------------ ...((((((.((.....)).))(-((((((.....)))))))...))))..(((----------(..((((((((...)))))))))))--).....-.......------------ ( -29.20, z-score = -1.58, R) >droVir3.scaffold_13047 8519262 91 + 19223366 UAACUCGGCGCAAUUAGUGGGCG-CCAUAAUUCGUUUGCGGCCAGCGAACGGAG----------CGCAGGAUAUGGCGCAUAUCCUGCU--CAUAAA-UAUUAAC------------ ...((((((((.........)))-))......(((((((.....))))))))))----------.((((((((((...)))))))))).--......-.......------------ ( -36.30, z-score = -3.47, R) >droGri2.scaffold_14624 3769339 95 + 4233967 UAACUCAGCGCAAUUAGUGGGCG-CCAUAAUUCGUUUGCGGCCGAGGCGAAACGGA---G----CAAAGGAUAUGCCGCGUAUCCUGGCACCAUAAA-UA-CUAC------------ ............(((.((((..(-(((.......(((((..(((........))).---)----))))(((((((...))))))))))).)))).))-).-....------------ ( -26.60, z-score = -0.11, R) >consensus UAACUUGGUGCAAUUAGUGGGCGGCCAUAAUUCGUUUGCGGCCAGACUUCUGAGGG___G____AGCAGGAUAUGCCGCAUAUCCUGGC_UCAUAAA_UAAAGACCUGCAAGCACAC ...((.(.(((((..(((.((...))...)))...))))).).))....................((((((((((...))))))))))............................. (-13.95 = -13.67 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:01:58 2011