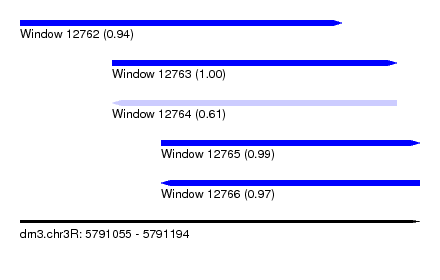
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 5,791,055 – 5,791,194 |
| Length | 139 |
| Max. P | 0.995892 |

| Location | 5,791,055 – 5,791,167 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 77.30 |
| Shannon entropy | 0.44291 |
| G+C content | 0.46992 |
| Mean single sequence MFE | -19.41 |
| Consensus MFE | -13.74 |
| Energy contribution | -14.10 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.936143 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
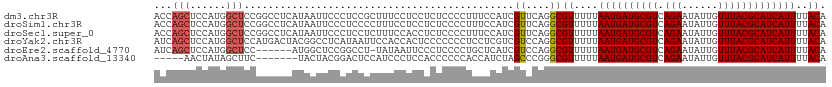
>dm3.chr3R 5791055 112 + 27905053 ACCAGCUCCAUGGCUCCGGCCUCAUAAUUCCCUCCGCUUUCCUCCUCUCCCCUUUCCAUCGUUCAGGCGUUUUUAAUGAUGCGUCAGAAUAUUGUUUACGCAUCAUUUUACA ...........(((....)))..............((((.........................))))((....((((((((((.(((......)))))))))))))..)). ( -18.01, z-score = -1.17, R) >droSim1.chr3R 11942626 112 - 27517382 ACCAGCUCCAUGGCUCCGGCCUCAUAAUUCCCUCCCCUUUCCUCCUCUCCCCUUUCCAUCGUUCAGGCGUUUUUAAUGAUGCGUCAGAAUAUUGUUUACGCAUCAUUUUACA .(((......))).....((((..........................................))))((....((((((((((.(((......)))))))))))))..)). ( -17.75, z-score = -1.60, R) >droSec1.super_0 4993860 112 - 21120651 ACCAGCUCCAUGGCUCCGGCCUCAUAAUUCCCUCCUCUUUCCACCUCUCCCCUUUCCAUCGUUCAGGCGUUUUUAAUGAUGCGUCAGAAUAUUGUUUACGCAUCAUUUUACA .(((......))).....((((..........................................))))((....((((((((((.(((......)))))))))))))..)). ( -17.75, z-score = -1.54, R) >droYak2.chr3R 9838897 112 + 28832112 AUCAGCUCCAUGGCUCCAUGACUACGGCCUCAUAAUUCCACCACUCCCCCCCUCCUCGUCGUCCAGGCGUUUUUAAUGAUGCGUCAGAAUAUUGUUUACGCAUCAUUUUACA ....(((.((((....))))...(((((.............................)))))...)))((....((((((((((.(((......)))))))))))))..)). ( -21.55, z-score = -1.65, R) >droEre2.scaffold_4770 1935088 105 - 17746568 AUCAGCUCCAUGGCUCC------AUGGCUCCGGCCU-UAUAAUUCCCUCCCCUGCUCAUCGUCCAGGCGUUUUUAAUGAUGCGUCAGAAUAUUGUUUACGCAUCAUUUUACA ....((((((((....)------))))....)))..-.............((((.........)))).((....((((((((((.(((......)))))))))))))..)). ( -24.00, z-score = -1.66, R) >droAna3.scaffold_13340 6351216 100 + 23697760 -----AACUAUAGCUUC-------UACUACGGACUCCAUCCCUCCACCCCCCACCAUCUAGCCCGGGCGUUUUUAAUGAUGCGUCAGAAUAUUGUUUACGCAUCAUUUUACA -----..(..(((....-------..)))..)............................((....))((....((((((((((.(((......)))))))))))))..)). ( -17.40, z-score = -1.53, R) >consensus ACCAGCUCCAUGGCUCCGGCCUCAUAAUUCCCUCCUCUUUCCUCCUCUCCCCUUCCCAUCGUCCAGGCGUUUUUAAUGAUGCGUCAGAAUAUUGUUUACGCAUCAUUUUACA ...(((......))).............................................((....))((....((((((((((.(((......)))))))))))))..)). (-13.74 = -14.10 + 0.36)
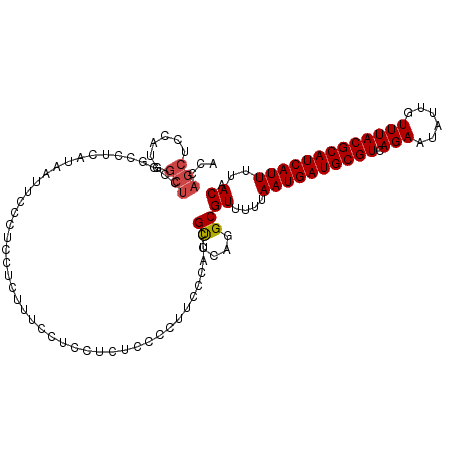
| Location | 5,791,087 – 5,791,186 |
|---|---|
| Length | 99 |
| Sequences | 9 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 75.97 |
| Shannon entropy | 0.45825 |
| G+C content | 0.44801 |
| Mean single sequence MFE | -26.24 |
| Consensus MFE | -18.81 |
| Energy contribution | -18.49 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.86 |
| SVM RNA-class probability | 0.995892 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 5791087 99 + 27905053 UCCGCUUUCCUCCUCUCCCCUUUCCAUCGUUCAGGCGUUUUUAAUGAUGCGUCAGAAUAUUGUUUACGCAUCAUUUUACAUGUCGGCAAAUGGGAAUGG----------------- .................((.(((((((......(((((.(..((((((((((.(((......)))))))))))))..).))))).....))))))).))----------------- ( -26.00, z-score = -2.56, R) >droSim1.chr3R 11942658 99 - 27517382 UCCCCUUUCCUCCUCUCCCCUUUCCAUCGUUCAGGCGUUUUUAAUGAUGCGUCAGAAUAUUGUUUACGCAUCAUUUUACAUGUCGGCAAAUGGGAAUGG----------------- .................((.(((((((......(((((.(..((((((((((.(((......)))))))))))))..).))))).....))))))).))----------------- ( -26.00, z-score = -2.88, R) >droSec1.super_0 4993892 99 - 21120651 UCCUCUUUCCACCUCUCCCCUUUCCAUCGUUCAGGCGUUUUUAAUGAUGCGUCAGAAUAUUGUUUACGCAUCAUUUUACAUGUCGGCAAAUGGGAAUGG----------------- .................((.(((((((......(((((.(..((((((((((.(((......)))))))))))))..).))))).....))))))).))----------------- ( -26.00, z-score = -2.93, R) >droYak2.chr3R 9838937 108 + 28832112 --------CCACUCCCCCCCUCCUCGUCGUCCAGGCGUUUUUAAUGAUGCGUCAGAAUAUUGUUUACGCAUCAUUUUACAUGUCGGCAAAUGGGAAUUGGAAUGGGACUGGGCUGC --------(((.((((..(((((.(((...((.(((((.(..((((((((((.(((......)))))))))))))..).)))))))...))))))...))...)))).)))..... ( -34.50, z-score = -1.48, R) >droEre2.scaffold_4770 1935126 86 - 17746568 -------------CCUCCCCUGCUCAUCGUCCAGGCGUUUUUAAUGAUGCGUCAGAAUAUUGUUUACGCAUCAUUUUACAUGUCGGCAAAUGGGAAUGG----------------- -------------.....((..(((((...((.(((((.(..((((((((((.(((......)))))))))))))..).)))))))...)))))...))----------------- ( -25.10, z-score = -2.23, R) >droAna3.scaffold_13340 6351240 95 + 23697760 ----CAUCCCUCCACCCCCCACCAUCUAGCCCGGGCGUUUUUAAUGAUGCGUCAGAAUAUUGUUUACGCAUCAUUUUACAUGUCGGCAAAUGGGCAUGG----------------- ----.......(((...((((.......(((..(((((.(..((((((((((.(((......)))))))))))))..).))))))))...))))..)))----------------- ( -25.90, z-score = -1.52, R) >dp4.chr2 25610683 75 - 30794189 ------------------------CCUCGUUCAGGCGUUUUUAAUGAUGCGUCAGAAUAUUGUUUACGCAUCAUUUUACAUGUCGGCAAAUGGGAAUGG----------------- ------------------------.((((((..(((((.(..((((((((((.(((......)))))))))))))..).)))))....)))))).....----------------- ( -22.10, z-score = -2.82, R) >droPer1.super_19 975968 75 - 1869541 ------------------------CCUCGUUCAGGCGUUUUUAAUGAUGCGUCAGAAUAUUGUUUACGCAUCAUUUUACAUGUCGGCAAAUGGGAAUGG----------------- ------------------------.((((((..(((((.(..((((((((((.(((......)))))))))))))..).)))))....)))))).....----------------- ( -22.10, z-score = -2.82, R) >droWil1.scaffold_181130 387839 106 - 16660200 ---------CUCCUCCACAAUUUGCCUCGUUCAGGCGUUUUUAAUGAUGCGUCAGAAUAUUGUUUACGCAUCAUUUUACAUGUCGGCAAACGGCUACAGAGAGUGACAGAGACAA- ---------(((...(((..((((..(((((..(((((.(..((((((((((.(((......)))))))))))))..).)))))....)))))...))))..)))...)))....- ( -28.50, z-score = -2.00, R) >consensus ________CCUCCUCUCCCCUUUCCAUCGUUCAGGCGUUUUUAAUGAUGCGUCAGAAUAUUGUUUACGCAUCAUUUUACAUGUCGGCAAAUGGGAAUGG_________________ ............................(((..(((((.(..((((((((((.(((......)))))))))))))..).))))))))............................. (-18.81 = -18.49 + -0.32)


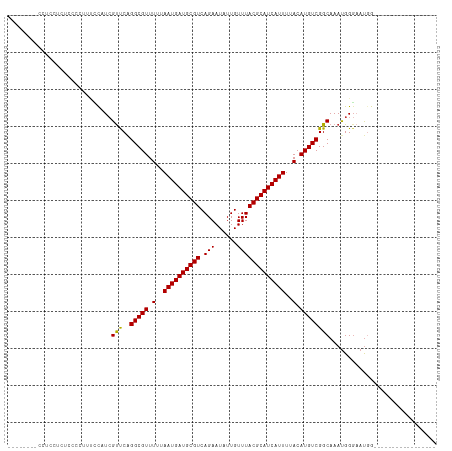
| Location | 5,791,087 – 5,791,186 |
|---|---|
| Length | 99 |
| Sequences | 9 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 75.97 |
| Shannon entropy | 0.45825 |
| G+C content | 0.44801 |
| Mean single sequence MFE | -23.82 |
| Consensus MFE | -12.32 |
| Energy contribution | -12.32 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.614585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 5791087 99 - 27905053 -----------------CCAUUCCCAUUUGCCGACAUGUAAAAUGAUGCGUAAACAAUAUUCUGACGCAUCAUUAAAAACGCCUGAACGAUGGAAAGGGGAGAGGAGGAAAGCGGA -----------------((.(((((.(((.(((........((((((((((.............)))))))))).....((......)).)))))).))))).))........... ( -24.62, z-score = -1.51, R) >droSim1.chr3R 11942658 99 + 27517382 -----------------CCAUUCCCAUUUGCCGACAUGUAAAAUGAUGCGUAAACAAUAUUCUGACGCAUCAUUAAAAACGCCUGAACGAUGGAAAGGGGAGAGGAGGAAAGGGGA -----------------....((((.(((.((..(......((((((((((.............)))))))))).....(.(((...........))).)...)..))))).)))) ( -25.82, z-score = -1.72, R) >droSec1.super_0 4993892 99 + 21120651 -----------------CCAUUCCCAUUUGCCGACAUGUAAAAUGAUGCGUAAACAAUAUUCUGACGCAUCAUUAAAAACGCCUGAACGAUGGAAAGGGGAGAGGUGGAAAGAGGA -----------------((((((((.(((.(((........((((((((((.............)))))))))).....((......)).)))))).))))...))))........ ( -24.42, z-score = -1.48, R) >droYak2.chr3R 9838937 108 - 28832112 GCAGCCCAGUCCCAUUCCAAUUCCCAUUUGCCGACAUGUAAAAUGAUGCGUAAACAAUAUUCUGACGCAUCAUUAAAAACGCCUGGACGACGAGGAGGGGGGGAGUGG-------- .....(((.((((.((((...(((...((((((...(((..((((((((((.............))))))))))....)))..))).)))...))))))))))).)))-------- ( -32.02, z-score = -1.22, R) >droEre2.scaffold_4770 1935126 86 + 17746568 -----------------CCAUUCCCAUUUGCCGACAUGUAAAAUGAUGCGUAAACAAUAUUCUGACGCAUCAUUAAAAACGCCUGGACGAUGAGCAGGGGAGG------------- -----------------...(((((...(((((........((((((((((.............)))))))))).....((......)).)).))).))))).------------- ( -22.02, z-score = -1.60, R) >droAna3.scaffold_13340 6351240 95 - 23697760 -----------------CCAUGCCCAUUUGCCGACAUGUAAAAUGAUGCGUAAACAAUAUUCUGACGCAUCAUUAAAAACGCCCGGGCUAGAUGGUGGGGGGUGGAGGGAUG---- -----------------.(((.(((.((..((.........((((((((((.............)))))))))).......(((.(.(.....).).)))))..))))))))---- ( -31.12, z-score = -1.61, R) >dp4.chr2 25610683 75 + 30794189 -----------------CCAUUCCCAUUUGCCGACAUGUAAAAUGAUGCGUAAACAAUAUUCUGACGCAUCAUUAAAAACGCCUGAACGAGG------------------------ -----------------......((..((((......))))((((((((((.............))))))))))................))------------------------ ( -13.22, z-score = -1.38, R) >droPer1.super_19 975968 75 + 1869541 -----------------CCAUUCCCAUUUGCCGACAUGUAAAAUGAUGCGUAAACAAUAUUCUGACGCAUCAUUAAAAACGCCUGAACGAGG------------------------ -----------------......((..((((......))))((((((((((.............))))))))))................))------------------------ ( -13.22, z-score = -1.38, R) >droWil1.scaffold_181130 387839 106 + 16660200 -UUGUCUCUGUCACUCUCUGUAGCCGUUUGCCGACAUGUAAAAUGAUGCGUAAACAAUAUUCUGACGCAUCAUUAAAAACGCCUGAACGAGGCAAAUUGUGGAGGAG--------- -..............((((...((.(((((((.........((((((((((.............)))))))))).....((......)).))))))).))...))))--------- ( -27.92, z-score = -1.74, R) >consensus _________________CCAUUCCCAUUUGCCGACAUGUAAAAUGAUGCGUAAACAAUAUUCUGACGCAUCAUUAAAAACGCCUGAACGAGGGAAAGGGGAGAGGAGG________ ...........................((((......))))((((((((((.............)))))))))).......................................... (-12.32 = -12.32 + 0.00)

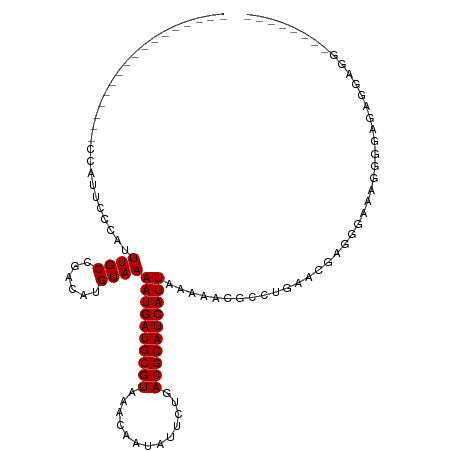
| Location | 5,791,104 – 5,791,194 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 75.22 |
| Shannon entropy | 0.52081 |
| G+C content | 0.44256 |
| Mean single sequence MFE | -26.48 |
| Consensus MFE | -17.75 |
| Energy contribution | -17.59 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.992920 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
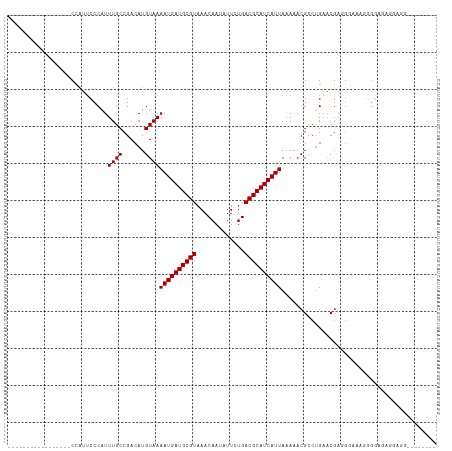
>dm3.chr3R 5791104 90 + 27905053 CCCUUUCCAUCGUUCAGGCGUUUUUAAUGAUGCGUCAGAAUAUUGUUUACGCAUCAUUUUACAUGUCGGCAAAUGGGAAU------------GGGCUGCAGU------ ((((((((((......(((((.(..((((((((((.(((......)))))))))))))..).))))).....))))))).------------))).......------ ( -28.80, z-score = -2.86, R) >droSim1.chr3R 11942675 90 - 27517382 CCCUUUCCAUCGUUCAGGCGUUUUUAAUGAUGCGUCAGAAUAUUGUUUACGCAUCAUUUUACAUGUCGGCAAAUGGGAAU------------GGGCUGCAGU------ ((((((((((......(((((.(..((((((((((.(((......)))))))))))))..).))))).....))))))).------------))).......------ ( -28.80, z-score = -2.86, R) >droSec1.super_0 4993909 90 - 21120651 CCCUUUCCAUCGUUCAGGCGUUUUUAAUGAUGCGUCAGAAUAUUGUUUACGCAUCAUUUUACAUGUCGGCAAAUGGGAAU------------GGGCUGCAGU------ ((((((((((......(((((.(..((((((((((.(((......)))))))))))))..).))))).....))))))).------------))).......------ ( -28.80, z-score = -2.86, R) >droYak2.chr3R 9838946 108 + 28832112 CCCUCCUCGUCGUCCAGGCGUUUUUAAUGAUGCGUCAGAAUAUUGUUUACGCAUCAUUUUACAUGUCGGCAAAUGGGAAUUGGAAUGGGACUGGGCUGCAGUUGCAGU ((((((...((.(((.(((((.(..((((((((((.(((......)))))))))))))..).))))))).....).))...)))..))).....(((((....))))) ( -32.20, z-score = -0.93, R) >droEre2.scaffold_4770 1935130 90 - 17746568 CCCUGCUCAUCGUCCAGGCGUUUUUAAUGAUGCGUCAGAAUAUUGUUUACGCAUCAUUUUACAUGUCGGCAAAUGGGAAU------------GGGCUGCAGU------ (((..(((((...((.(((((.(..((((((((((.(((......)))))))))))))..).)))))))...)))))...------------))).......------ ( -28.50, z-score = -2.03, R) >droAna3.scaffold_13340 6351253 91 + 23697760 CCCACCAUCUAGCCCGGGCGUUUUUAAUGAUGCGUCAGAAUAUUGUUUACGCAUCAUUUUACAUGUCGGCAAAUGGGCAU------------GGGCUGUGAGU----- .......((((((((((((((.(..((((((((((.(((......)))))))))))))..).))))).((......)).)------------)))))).))..----- ( -30.60, z-score = -2.05, R) >dp4.chr2 25610683 84 - 30794189 -------CCUCGUUCAGGCGUUUUUAAUGAUGCGUCAGAAUAUUGUUUACGCAUCAUUUUACAUGUCGGCAAAUGGGAAU------------GGGUUGUGGAG----- -------.(((((((.(((((.(..((((((((((.(((......)))))))))))))..).))))).........))))------------)))........----- ( -23.30, z-score = -1.94, R) >droPer1.super_19 975968 89 - 1869541 -------CCUCGUUCAGGCGUUUUUAAUGAUGCGUCAGAAUAUUGUUUACGCAUCAUUUUACAUGUCGGCAAAUGGGAAU------------GGGUUGUGGAGUGGAG -------.(((((((.(((((.(..((((((((((.(((......)))))))))))))..).))))).........))))------------)))............. ( -23.30, z-score = -1.28, R) >droWil1.scaffold_181130 387847 107 - 16660200 CAAUUUGCCUCGUUCAGGCGUUUUUAAUGAUGCGUCAGAAUAUUGUUUACGCAUCAUUUUACAUGUCGGCAAACGG-CUACAGAGAGUGACAGAGACAAAGCAAGAGA ...((((.(((..((((((((.(..((((((((((.(((......)))))))))))))..).)))))(((.....)-))........)))..))).))))........ ( -27.00, z-score = -1.30, R) >droVir3.scaffold_13047 8501043 78 - 19223366 -------CCUUGUUCAGGCGUUUU-AAUGAUGCGUCAGAAUAUUGUUUACGCAUCAUUUUACAUGUCGGCAAAUGAGGACCACAGC---------------------- -------(((..((..(((((...-((((((((((.(((......)))))))))))))....)))))....))..)))........---------------------- ( -22.70, z-score = -2.66, R) >droMoj3.scaffold_6540 31072137 80 + 34148556 -------CCUUGUUCAGGCGUUUU-AAUGAUGCGUCAGAAUAUUGUUUACGCAUCAUUUUACAUGUCGGCAAAUGGGCUACGCAGCGC-------------------- -------((((((...(((((...-((((((((((.(((......)))))))))))))....))))).))))..))(((....)))..-------------------- ( -21.80, z-score = -1.17, R) >droGri2.scaffold_14624 3747270 80 - 4233967 ---------CCUCUCAGGCGUUUU-AAUGAUGCGUCAGAAUAUUGUUUACGCAUCACUUUACAUGUCGGCAAACCGAUGA------------GGACGAUGGU------ ---------((((((.(((((...-((((((((((.(((......))))))))))).))...)))))((....)))).))------------))........------ ( -22.00, z-score = -1.44, R) >consensus CCCU___CCUCGUUCAGGCGUUUUUAAUGAUGCGUCAGAAUAUUGUUUACGCAUCAUUUUACAUGUCGGCAAAUGGGAAU____________GGGCUGCAGU______ ...........(((..(((((....((((((((((.(((......)))))))))))))....))))))))...................................... (-17.75 = -17.59 + -0.16)

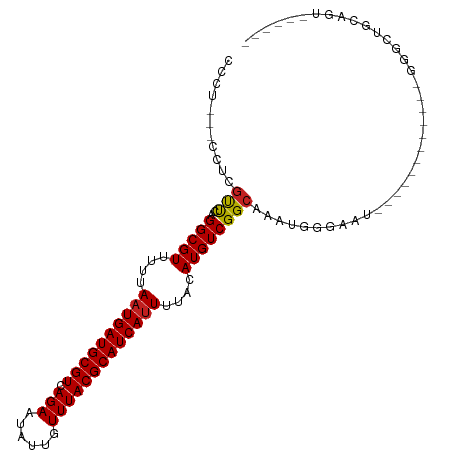
| Location | 5,791,104 – 5,791,194 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 75.22 |
| Shannon entropy | 0.52081 |
| G+C content | 0.44256 |
| Mean single sequence MFE | -20.61 |
| Consensus MFE | -12.22 |
| Energy contribution | -12.14 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.970837 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 5791104 90 - 27905053 ------ACUGCAGCCC------------AUUCCCAUUUGCCGACAUGUAAAAUGAUGCGUAAACAAUAUUCUGACGCAUCAUUAAAAACGCCUGAACGAUGGAAAGGG ------.......(((------------.((((...((((......))))((((((((((.............)))))))))).................)))).))) ( -20.82, z-score = -1.79, R) >droSim1.chr3R 11942675 90 + 27517382 ------ACUGCAGCCC------------AUUCCCAUUUGCCGACAUGUAAAAUGAUGCGUAAACAAUAUUCUGACGCAUCAUUAAAAACGCCUGAACGAUGGAAAGGG ------.......(((------------.((((...((((......))))((((((((((.............)))))))))).................)))).))) ( -20.82, z-score = -1.79, R) >droSec1.super_0 4993909 90 + 21120651 ------ACUGCAGCCC------------AUUCCCAUUUGCCGACAUGUAAAAUGAUGCGUAAACAAUAUUCUGACGCAUCAUUAAAAACGCCUGAACGAUGGAAAGGG ------.......(((------------.((((...((((......))))((((((((((.............)))))))))).................)))).))) ( -20.82, z-score = -1.79, R) >droYak2.chr3R 9838946 108 - 28832112 ACUGCAACUGCAGCCCAGUCCCAUUCCAAUUCCCAUUUGCCGACAUGUAAAAUGAUGCGUAAACAAUAUUCUGACGCAUCAUUAAAAACGCCUGGACGACGAGGAGGG .((((....))))((..(((....((((........((((......))))((((((((((.............)))))))))).........)))).)))..)).... ( -25.62, z-score = -1.35, R) >droEre2.scaffold_4770 1935130 90 + 17746568 ------ACUGCAGCCC------------AUUCCCAUUUGCCGACAUGUAAAAUGAUGCGUAAACAAUAUUCUGACGCAUCAUUAAAAACGCCUGGACGAUGAGCAGGG ------.((((....(------------(((.(((.((((......))))((((((((((.............)))))))))).........)))..)))).)))).. ( -20.92, z-score = -1.12, R) >droAna3.scaffold_13340 6351253 91 - 23697760 -----ACUCACAGCCC------------AUGCCCAUUUGCCGACAUGUAAAAUGAUGCGUAAACAAUAUUCUGACGCAUCAUUAAAAACGCCCGGGCUAGAUGGUGGG -----.(((((.....------------..((((..((((......))))((((((((((.............))))))))))..........))))......))))) ( -26.64, z-score = -2.06, R) >dp4.chr2 25610683 84 + 30794189 -----CUCCACAACCC------------AUUCCCAUUUGCCGACAUGUAAAAUGAUGCGUAAACAAUAUUCUGACGCAUCAUUAAAAACGCCUGAACGAGG------- -----(((........------------........((((......))))((((((((((.............))))))))))..............))).------- ( -14.62, z-score = -1.97, R) >droPer1.super_19 975968 89 + 1869541 CUCCACUCCACAACCC------------AUUCCCAUUUGCCGACAUGUAAAAUGAUGCGUAAACAAUAUUCUGACGCAUCAUUAAAAACGCCUGAACGAGG------- .....(((........------------........((((......))))((((((((((.............))))))))))..............))).------- ( -15.12, z-score = -2.15, R) >droWil1.scaffold_181130 387847 107 + 16660200 UCUCUUGCUUUGUCUCUGUCACUCUCUGUAG-CCGUUUGCCGACAUGUAAAAUGAUGCGUAAACAAUAUUCUGACGCAUCAUUAAAAACGCCUGAACGAGGCAAAUUG ....(((((((((.((((((((.....)).(-(.....)).)))).....((((((((((.............))))))))))..........))))))))))).... ( -25.52, z-score = -2.43, R) >droVir3.scaffold_13047 8501043 78 + 19223366 ----------------------GCUGUGGUCCUCAUUUGCCGACAUGUAAAAUGAUGCGUAAACAAUAUUCUGACGCAUCAUU-AAAACGCCUGAACAAGG------- ----------------------((((((((........))).))).))..((((((((((.............))))))))))-......(((.....)))------- ( -17.22, z-score = -1.43, R) >droMoj3.scaffold_6540 31072137 80 - 34148556 --------------------GCGCUGCGUAGCCCAUUUGCCGACAUGUAAAAUGAUGCGUAAACAAUAUUCUGACGCAUCAUU-AAAACGCCUGAACAAGG------- --------------------.....((((.......((((......))))((((((((((.............))))))))))-...))))..........------- ( -17.02, z-score = -0.61, R) >droGri2.scaffold_14624 3747270 80 + 4233967 ------ACCAUCGUCC------------UCAUCGGUUUGCCGACAUGUAAAGUGAUGCGUAAACAAUAUUCUGACGCAUCAUU-AAAACGCCUGAGAGG--------- ------........((------------((.((((....)))).......((((((((((.............))))))))))-...........))))--------- ( -22.22, z-score = -2.46, R) >consensus ______ACUGCAGCCC____________AUUCCCAUUUGCCGACAUGUAAAAUGAUGCGUAAACAAUAUUCUGACGCAUCAUUAAAAACGCCUGAACGAGG___AGGG ....................................((((......))))((((((((((.............))))))))))......................... (-12.22 = -12.14 + -0.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:01:57 2011