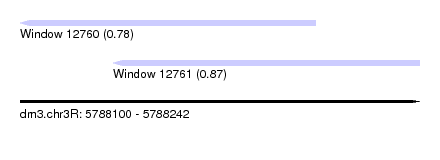
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 5,788,100 – 5,788,242 |
| Length | 142 |
| Max. P | 0.872148 |

| Location | 5,788,100 – 5,788,205 |
|---|---|
| Length | 105 |
| Sequences | 8 |
| Columns | 124 |
| Reading direction | reverse |
| Mean pairwise identity | 75.32 |
| Shannon entropy | 0.42812 |
| G+C content | 0.33205 |
| Mean single sequence MFE | -23.38 |
| Consensus MFE | -12.87 |
| Energy contribution | -12.78 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.775713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
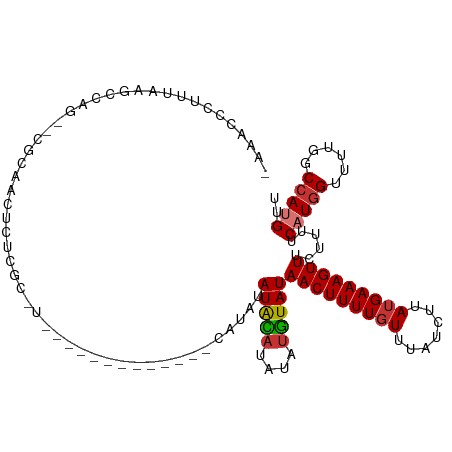
>dm3.chr3R 5788100 105 - 27905053 ------CGUGCAUAUAUGCAUA------UAUGUAUAACUUUUGUUUAUCUUAUGAAAGUUUCUUUUCAUAGUUUUGGCCAUGUUUGCCAGCCAACA-------GGCAGCAGGAGAAACAAAAGC ------.(((((((((....))------)))))))..(((((((((.(((((((((((....))))))))(((.((((.......))))(((....-------)))))).))).))))))))). ( -30.40, z-score = -3.10, R) >droSim1.chr3R 11939691 96 + 27517382 ---------------AUGCAUA------UAUGUAUAACUUUUGUUUAUCUUAUGAAAGUUUCUUUUCAUGGUUUUGGCCAUGUUUGCCAGCCAACA-------GGCAGCAGGAGAAACAAAAGC ---------------(((((..------..)))))..(((((((((.((((...............((((((....))))))...((..(((....-------))).)))))).))))))))). ( -25.10, z-score = -1.74, R) >droYak2.chr3R 9835771 112 - 28832112 ------UAUGCAUUUAUGCAUA------UAUGUAUAACUUUUGUUUAUCUUAUGAAAGUUUCUUUUCAUGGUUUUGGCCAUGUUUGCCAGCCAACAGGCAGCAGGCAGCAGGAAAAACAAAAGC ------((((((....))))))------.........(((((((((.(((((((((((....)))))))(((....))).(((((((..(((....))).)))))))..)))).))))))))). ( -33.40, z-score = -2.68, R) >droEre2.scaffold_4770 1932033 111 + 17746568 ------CAUGCAUAUAUGCAUAUACUCGUAUGUAUAACUUUUGUUUAUCUUAUGAAAGUUUCUUUUCAUGGUUUUGGCCAUGUUUGCCAGCCAACA-------GGCAGCAGGAAAAACAAAAGC ------........(((((((((....))))))))).(((((((((.((((...............((((((....))))))...((..(((....-------))).)))))).))))))))). ( -30.60, z-score = -2.61, R) >droAna3.scaffold_13340 6348230 104 - 23697760 ACAUGCCCCGUGCAUAUAUAUG------CAUAUAUAACUUUUGUUUAUCUUAUGAAAGUUUCUCUUCAUGGCAUUUUCCAUGUUUGCCACAGU------------CGACAAGAAAAACAAAA-- .........((((((....)))------)))........(((((((.(((((((((........)))))((((..(.....)..)))).....------------....)))).))))))).-- ( -20.40, z-score = -1.97, R) >dp4.chr2 25607880 90 + 30794189 -----------AUACAUAUAUG------CAUAUAUAACUUUUUUUUAUCUUAUGAAAGUUUCCUUUC-UGGCAUUUUCCAUGUUUG-CAGGCA------------CAAAAAAAAAAACAAA--- -----------.........((------((.((((..................(((((....)))))-.((......)))))).))-))....------------................--- ( -9.10, z-score = 0.43, R) >droPer1.super_19 973122 88 + 1869541 -----------AUACAUAUAUG------CAUAUAUAACUUUUGUUUAUCUUAUGAAAGUUUCCUUUC-UGGCAUUUUCCAUGUUUG-CAGGCA------------CAAAAAAAUAAAAA----- -----------.........((------((.((((((((((..(.......)..)))))).((....-.))........)))).))-))....------------..............----- ( -10.50, z-score = 0.31, R) >droSec1.super_0 4990931 100 + 21120651 -----------AUGCAUAUACA------UAUGUAUAACUUUUGUUUAUCUUAUGAAAGUUUCUUUUCAUGGUUUUGGCCAUGUUUGCCAGCCAACA-------GGCAGCAGGAAAAACAAAAGC -----------(((((((....------)))))))..(((((((((.((((...............((((((....))))))...((..(((....-------))).)))))).))))))))). ( -27.50, z-score = -2.55, R) >consensus ___________AUAUAUACAUA______UAUGUAUAACUUUUGUUUAUCUUAUGAAAGUUUCUUUUCAUGGUUUUGGCCAUGUUUGCCAGCCAACA_______GGCAGCAGGAAAAACAAAAGC ...............(((((((......)))))))...((((((((.((((...............(((((......))))).((((.................)))).)))).)))))))).. (-12.87 = -12.78 + -0.09)
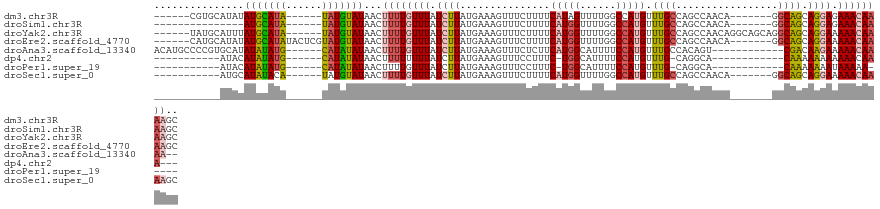
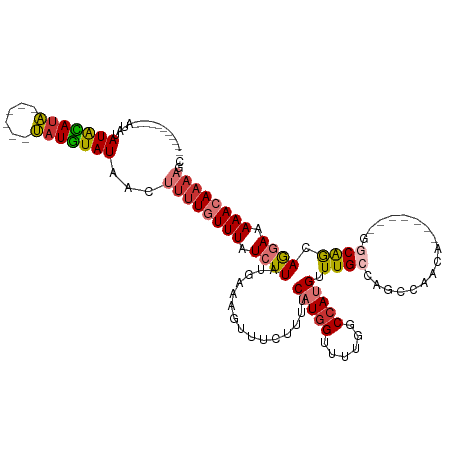
| Location | 5,788,133 – 5,788,242 |
|---|---|
| Length | 109 |
| Sequences | 8 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 71.20 |
| Shannon entropy | 0.55713 |
| G+C content | 0.35376 |
| Mean single sequence MFE | -21.47 |
| Consensus MFE | -8.60 |
| Energy contribution | -8.51 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.872148 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 5788133 109 - 27905053 -AAACCCUUCAGGCCAG--CGAAACUCUCGCAUAUGUG-GC-CGUGCAUAUAUGCAUAUAUGUAUAACUUUUGUUUAUCUUAUGAAAGUUUCUUUUCAUAGUUUUGGCCAUGUU -..........(((((.--..((((....(((((((((-..-....)))))))))...((((...((((((..(.......)..))))))......)))))))))))))..... ( -28.40, z-score = -2.85, R) >droSim1.chr3R 11939724 95 + 27517382 -AAACCCUUCAGGCCAG--GCCAACUCUCGC----------------AUAUAUGCAUAUAUGUAUAACUUUUGUUUAUCUUAUGAAAGUUUCUUUUCAUGGUUUUGGCCAUGUU -..........((((((--((((......((----------------(((((....)))))))..((((((..(.......)..))))))........))).)))))))..... ( -24.00, z-score = -2.98, R) >droYak2.chr3R 9835811 107 - 28832112 -AAACCCUUUAAGCCAG----CAACUCUGGCAUAUGUG-GC-UAUGCAUUUAUGCAUAUAUGUAUAACUUUUGUUUAUCUUAUGAAAGUUUCUUUUCAUGGUUUUGGCCAUGUU -...........(((((----.....)))))..(((((-((-((.((..(((((((....))))))).............((((((((....))))))))))..))))))))). ( -26.80, z-score = -2.11, R) >droEre2.scaffold_4770 1932066 113 + 17746568 -AAACCCUUUAAGCCAGCAACUCUCGCAUAUGUGGCCAUGCAUAUAUGCAUAUACUCGUAUGUAUAACUUUUGUUUAUCUUAUGAAAGUUUCUUUUCAUGGUUUUGGCCAUGUU -...........(((((.(((....(((((((((......))))))))).(((((......)))))..............((((((((....))))))))))))))))...... ( -26.00, z-score = -2.12, R) >droAna3.scaffold_13340 6348256 111 - 23697760 CAAACCCUUUAAGCCAC--AUACUCAUAUGUGGCGACAUGC-CCCGUGCAUAUAUAUGCAUAUAUAACUUUUGUUUAUCUUAUGAAAGUUUCUCUUCAUGGCAUUUUCCAUGUU ............(((((--(((....))))))))...((((-(..((((((....))))))....((((((..(.......)..)))))).........))))).......... ( -27.60, z-score = -3.87, R) >dp4.chr2 25607904 98 + 30794189 AGGACCCUUUAAGCCCUUCCGCGCUACUCCU---------------CAUACAUAUAUGCAUAUAUAACUUUUUUUUAUCUUAUGAAAGUUUCCUUUC-UGGCAUUUUCCAUGUU .(((........((......))((((.....---------------.....(((((....)))))..................(((((....)))))-))))....)))..... ( -10.90, z-score = -0.23, R) >droPer1.super_19 973144 98 + 1869541 AGGACCCUUUAAGCCCUUCCGCCCUACUCCU---------------CAUACAUAUAUGCAUAUAUAACUUUUGUUUAUCUUAUGAAAGUUUCCUUUC-UGGCAUUUUCCAUGUU .(((........((......)).........---------------.........((((......((((((..(.......)..)))))).......-..))))..)))..... ( -10.56, z-score = -0.02, R) >droSec1.super_0 4990964 97 + 21120651 -AAACCCUUCAAGCCAG----CAACUCUCGCAUAU------------AUGCAUAUACAUAUGUAUAACUUUUGUUUAUCUUAUGAAAGUUUCUUUUCAUGGUUUUGGCCAUGUU -...........(((((----.(((.........(------------(((((((....))))))))..............((((((((....))))))))))))))))...... ( -17.50, z-score = -1.52, R) >consensus _AAACCCUUUAAGCCAG__CGCAACUCUCGC_U_____________CAUAUAUACAUAUAUGUAUAACUUUUGUUUAUCUUAUGAAAGUUUCUUUUCAUGGUUUUGGCCAUGUU ...................................................(((((....)))))(((((((((.......)))))))))......(((((......))))).. ( -8.60 = -8.51 + -0.09)

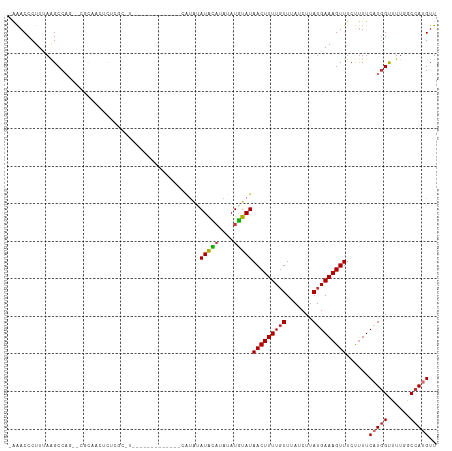
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:01:53 2011