
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 5,736,142 – 5,736,239 |
| Length | 97 |
| Max. P | 0.629784 |

| Location | 5,736,142 – 5,736,239 |
|---|---|
| Length | 97 |
| Sequences | 9 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 64.20 |
| Shannon entropy | 0.74414 |
| G+C content | 0.44167 |
| Mean single sequence MFE | -26.93 |
| Consensus MFE | -6.95 |
| Energy contribution | -7.14 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.73 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.26 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.629784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 5736142 97 + 27905053 CUUGAUCGAUGGUUACAAAGCCAUAAGCCAUCAUUAGCCAUGUGGCAAUUUAU-AUGCUGGCUUCACUUCGAUCUUA--UGAUCUCGAGAUCCGUUGUUU- ...(((((((((((....))))).((((((.(((..(((....))).......-))).))))))....))))))...--.((((....))))........- ( -25.90, z-score = -1.90, R) >droSim1.chr3R 11887398 97 - 27517382 CUUGAUCGAUGGUUGCACGGCCAUAAGCCAUCAUUAGCCAUGUGGCAAUUUAU-AUGCUGGCUUCACUUCGAUCUCA--AGAUCUCGAGAUCCGUUGUUU- ...(((((((((((....))))).((((((.(((..(((....))).......-))).))))))....))))))...--.((((....))))........- ( -26.70, z-score = -1.12, R) >droSec1.super_0 4938731 97 - 21120651 CUUGAUCGAUGGCUGCACGGCCAUAAGCCAUCAUUAGCCAUGUGGCAAUUUAU-AUGCUGGCUUCACUUCGAUCUCA--AGAUCUCGAGAUCCGUUGUUU- ...(((((((((((....))))).((((((.(((..(((....))).......-))).))))))....))))))...--.((((....))))........- ( -29.20, z-score = -1.75, R) >droEre2.scaffold_4770 1878845 97 - 17746568 CUUGAUCGAUGGUUCCACAGCCAUAAGCCAUCAUUAGCCAUGUGGCAAUUUAU-AUGCUGGCUUUACUCCGAUCUCA--UGAUCCCGCGAUCCGUUGUUU- ...((((((((((......)))))((((((.(((..(((....))).......-))).))))))......((((...--.))))...)))))........- ( -25.30, z-score = -1.50, R) >droYak2.chr3R 9781769 97 + 28832112 CUUGAUCGAUGGCUCCAAAGCCAUAAGCCAUCAUUAGCCAUGUGGCAAUUUAU-AUGCUGGCUUUACUUCGAUCUCA--UGAUCUCGCGAUCCGUUGUUU- ...(((((((((((....))))))((((((.(((..(((....))).......-))).))))))......((((...--.))))...)))))........- ( -28.10, z-score = -2.30, R) >droAna3.scaffold_13340 6296865 91 + 23697760 CUUGAUCGAUGGCUUCG-AGCCAUAAGCCAUCAUUAGCCAUGUGGCAAUUUAU-AU--UGCCAACUCUCUGGCACGA--UCGGGCC-AAAGCCGAAGC--- ..(((((((((((((..-......))))))))....((((..(((((((....-))--)))))......))))..))--)))(((.-...))).....--- ( -34.90, z-score = -3.53, R) >dp4.chr2 25555192 92 - 30794189 UUUGAUAGAUGGAUGCCGAACACUAAGUCAUCAUUAGCUCGAUACCCAUUUAUCAUGAUCAACGGACUCCGAUUUGGAUUCAGAUCGCGUUC--------- ..((((((((((.((.(((...((((.......)))).))).)).))))))))))........((((..(((((((....))))))).))))--------- ( -24.20, z-score = -1.74, R) >droPer1.super_19 920611 92 - 1869541 UUUGAUAGAUGGAUGCCGAGCACUAAGUCAUCAUUAGCUCGAUACCCAUUUAUCAUGAUCAACGGACUCUGAUUCGGAUUCAGAUCGCGUUC--------- ..((((((((((.((.(((((...............))))).)).)))))))))).....((((((.(((((.......))))))).)))).--------- ( -28.06, z-score = -2.46, R) >droVir3.scaffold_13047 8431284 99 - 19223366 CUUGAGGAGAAGGUAUUGAAGCCUGU-UUGUUAAGCUUUAAGCAUUAAGUUAAGUUAAUUGCAUUCAGCGCAUACGC-UUUAGCUUGCCAGAAGUUAUUGA (((((.(((.((((......)))).)-)).)))))((((..(((...((((((((.....))....((((....)))-))))))))))..))))....... ( -20.00, z-score = 0.92, R) >consensus CUUGAUCGAUGGCUGCACAGCCAUAAGCCAUCAUUAGCCAUGUGGCAAUUUAU_AUGCUGGCUUCACUCCGAUCUCA__UGAUCUCGCGAUCCGUUGUUU_ ...(((((((((((...........)))))))...........((((........))))...........))))........................... ( -6.95 = -7.14 + 0.19)

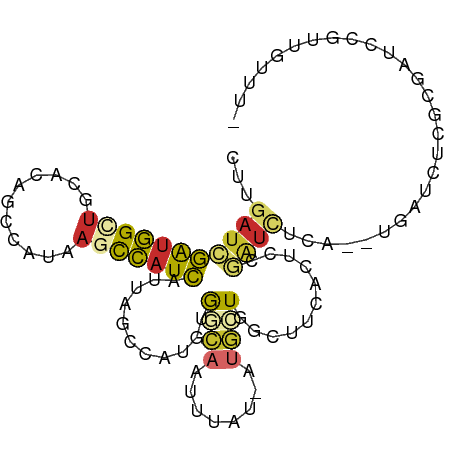

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:01:46 2011