
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 5,731,765 – 5,731,881 |
| Length | 116 |
| Max. P | 0.966094 |

| Location | 5,731,765 – 5,731,881 |
|---|---|
| Length | 116 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 82.07 |
| Shannon entropy | 0.35935 |
| G+C content | 0.31000 |
| Mean single sequence MFE | -19.19 |
| Consensus MFE | -11.00 |
| Energy contribution | -11.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.906519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
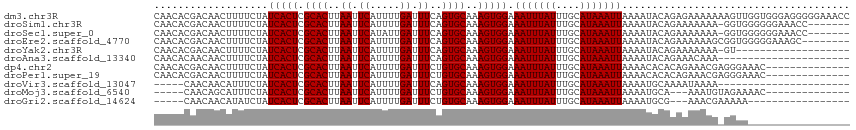
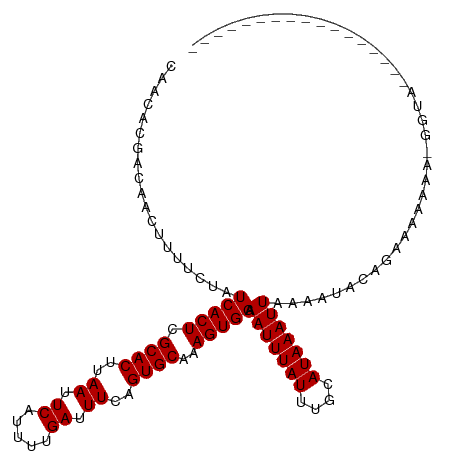
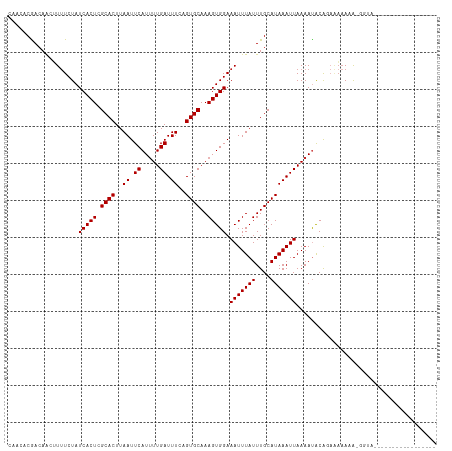
>dm3.chr3R 5731765 116 + 27905053 CAACACGACAACUUUUCUAUCACUCGCACUUAAUUCAUUUUGAUUUCAGUGCAAAGUGGAAAUUUAUUUGCAUAAAUUAAAAUACAGAGAAAAAAAGUUGGUGGGAGGGGGAAACC ...(((..((((((((((.(((((.(((((.((.((.....)).)).)))))..))))).(((((((....))))))).........)))...)))))))))).....((....)) ( -24.80, z-score = -2.00, R) >droSim1.chr3R 11882983 108 - 27517382 CAACACGACAACUUUUCUAUCACUCGCACUUAAUUCAUUUUGAUUUCAGUGCAAAGUGGAAAUUUAUUUGCAUAAAUUAAAAUACAGAAAAAAA-GGUGGGGGGAAACC------- .....(.((...((((((.(((((.(((((.((.((.....)).)).)))))..))))).(((((((....))))))).......))))))...-.)).).((....))------- ( -21.90, z-score = -2.35, R) >droSec1.super_0 4934339 108 - 21120651 CAACACGACAACUUUUCUAUCACUCGCACUUAAUUCAUAUUGAUUUCAGUGCAAAGUGGAAAUUUAUUUGCAUAAAUUAAAAUACAGAAAAAAA-GGUGGGGGGAAACC------- .....(.((...((((((.(((((.(((((.((.((.....)).)).)))))..))))).(((((((....))))))).......))))))...-.)).).((....))------- ( -21.90, z-score = -2.54, R) >droEre2.scaffold_4770 1874297 108 - 17746568 CAACACGACAACUUUUCUAUCACUCGCACUUAAUUCAUUUUGAUUUCAGUGCAAAGUGGAAAUUUAUUUGCAUAAAUUAAAAUACAGAAAAAAGCGGUGGGGGAAAGC-------- ...........(((((((.((((.(((.........((((((((((..(((((((...........)))))))))))))))))..........)))))))))))))).-------- ( -23.61, z-score = -2.79, R) >droYak2.chr3R 9777160 96 + 28832112 CAACACGACAACUUUUCUAUCACUCGCACUUAAUUCAUUUUGAUUUCAGUGCAAAGUGGAAAUUUAUUUGCAUAAAUUAAAAUACAGAAAAAAA-GU------------------- ............((((((.(((((.(((((.((.((.....)).)).)))))..))))).(((((((....))))))).......))))))...-..------------------- ( -16.50, z-score = -2.22, R) >droAna3.scaffold_13340 6292340 94 + 23697760 CAACACAACAACUUUUCUAUCACUCGCACUUAAUUCAUUUUGAUUUCAGUGCAAAGUGGAAAUUUAUUUGCAUAAAUUAAAAUACAGAAACAAA---------------------- .............(((((.(((((.(((((.((.((.....)).)).)))))..))))).(((((((....))))))).......)))))....---------------------- ( -15.40, z-score = -2.70, R) >dp4.chr2 25550540 102 - 30794189 CAACACGACAACUUUUCUAUCACUCGCACUUAAUUCAUUUUGAUUUCUGUGCAAAGUGGAAAUUUAUUUGCAUAAAUUAAAACACACAGAAACGAGGGAAAC-------------- .............(((((.((....((((..((.((.....)).))..))))...(((..(((((((....)))))))....)))........)).))))).-------------- ( -16.00, z-score = -1.44, R) >droPer1.super_19 915979 102 - 1869541 CAACACGACAACUUUUCUAUCACUCGCACUUAAUUCAUUUUGAUUUCUGUGCAAAGUGGAAAUUUAUUUGCAUAAAUUAAAACACACAGAAACGAGGGAAAC-------------- .............(((((.((....((((..((.((.....)).))..))))...(((..(((((((....)))))))....)))........)).))))).-------------- ( -16.00, z-score = -1.44, R) >droVir3.scaffold_13047 8426152 89 - 19223366 -----CAACAACAUUUCUAUCACUCGCACUUAAUUCAUUUUGAUUUCAGUGCAAAGUGGAAAUUUAUUUGCAUAAAUUAAAAUGCAAAAUAAAA---------------------- -----..............(((((.(((((.((.((.....)).)).)))))..)))))...(((((((((((........)))).))))))).---------------------- ( -18.20, z-score = -3.24, R) >droMoj3.scaffold_6540 30990791 94 + 34148556 -----CAACAGCAUUUCUAUCACUCGCACUUAAUUCAUUUUGAUUUCUGUGCAAAGUGGAAAUUUAUUUGCAUAAAUUAAAAUGCA---AAAUGUAGAAAAC-------------- -----........(((((((((((.((((..((.((.....)).))..))))..))))........(((((((........)))))---))..)))))))..-------------- ( -20.60, z-score = -2.51, R) >droGri2.scaffold_14624 3652331 91 - 4233967 -----CAACAACAUAUCUAUCACUCGCACUUAAUUCAUUUUGAUUUCUGUGCAAAGUGGAAAUUUAUUUGCAUAAAUUAAAAUGCG---AAACGAAAAA----------------- -----..............(((((.((((..((.((.....)).))..))))..))))).......(((((((........)))))---))........----------------- ( -16.20, z-score = -1.98, R) >consensus CAACACGACAACUUUUCUAUCACUCGCACUUAAUUCAUUUUGAUUUCAGUGCAAAGUGGAAAUUUAUUUGCAUAAAUUAAAAUACAGAAAAAAA_GGUA_________________ ...................(((((.((((..((.((.....)).))..))))..))))).(((((((....)))))))...................................... (-11.00 = -11.00 + 0.00)
| Location | 5,731,765 – 5,731,881 |
|---|---|
| Length | 116 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 82.07 |
| Shannon entropy | 0.35935 |
| G+C content | 0.31000 |
| Mean single sequence MFE | -21.19 |
| Consensus MFE | -13.63 |
| Energy contribution | -13.68 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.966094 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
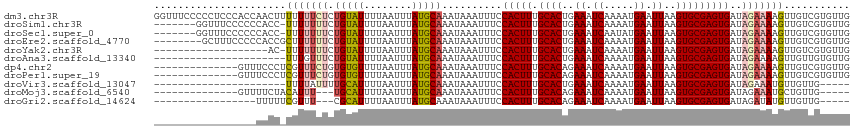
>dm3.chr3R 5731765 116 - 27905053 GGUUUCCCCCUCCCACCAACUUUUUUUCUCUGUAUUUUAAUUUAUGCAAAUAAAUUUCCACUUUGCACUGAAAUCAAAAUGAAUUAAGUGCGAGUGAUAGAAAAGUUGUCGUGUUG ((......))...(((((((((((((....(((((........)))))..........(((((.(((((.((.((.....)).)).))))))))))..))))))))))..)))... ( -24.50, z-score = -3.18, R) >droSim1.chr3R 11882983 108 + 27517382 -------GGUUUCCCCCCACC-UUUUUUUCUGUAUUUUAAUUUAUGCAAAUAAAUUUCCACUUUGCACUGAAAUCAAAAUGAAUUAAGUGCGAGUGAUAGAAAAGUUGUCGUGUUG -------..........((((-..(((((((((.....(((((((....)))))))..(((((.(((((.((.((.....)).)).)))))))))))))))))))..)..)))... ( -22.90, z-score = -2.96, R) >droSec1.super_0 4934339 108 + 21120651 -------GGUUUCCCCCCACC-UUUUUUUCUGUAUUUUAAUUUAUGCAAAUAAAUUUCCACUUUGCACUGAAAUCAAUAUGAAUUAAGUGCGAGUGAUAGAAAAGUUGUCGUGUUG -------..........((((-..(((((((((.....(((((((....)))))))..(((((.(((((.((.((.....)).)).)))))))))))))))))))..)..)))... ( -22.90, z-score = -2.99, R) >droEre2.scaffold_4770 1874297 108 + 17746568 --------GCUUUCCCCCACCGCUUUUUUCUGUAUUUUAAUUUAUGCAAAUAAAUUUCCACUUUGCACUGAAAUCAAAAUGAAUUAAGUGCGAGUGAUAGAAAAGUUGUCGUGUUG --------.........(((.((.(((((((((.....(((((((....)))))))..(((((.(((((.((.((.....)).)).)))))))))))))))))))..)).)))... ( -25.20, z-score = -3.81, R) >droYak2.chr3R 9777160 96 - 28832112 -------------------AC-UUUUUUUCUGUAUUUUAAUUUAUGCAAAUAAAUUUCCACUUUGCACUGAAAUCAAAAUGAAUUAAGUGCGAGUGAUAGAAAAGUUGUCGUGUUG -------------------((-..(((((((((.....(((((((....)))))))..(((((.(((((.((.((.....)).)).)))))))))))))))))))..))....... ( -22.30, z-score = -2.96, R) >droAna3.scaffold_13340 6292340 94 - 23697760 ----------------------UUUGUUUCUGUAUUUUAAUUUAUGCAAAUAAAUUUCCACUUUGCACUGAAAUCAAAAUGAAUUAAGUGCGAGUGAUAGAAAAGUUGUUGUGUUG ----------------------....(((((((.....(((((((....)))))))..(((((.(((((.((.((.....)).)).)))))))))))))))))............. ( -19.60, z-score = -2.09, R) >dp4.chr2 25550540 102 + 30794189 --------------GUUUCCCUCGUUUCUGUGUGUUUUAAUUUAUGCAAAUAAAUUUCCACUUUGCACAGAAAUCAAAAUGAAUUAAGUGCGAGUGAUAGAAAAGUUGUCGUGUUG --------------......(((((((((((((((...(((((((....)))))))...))...)))))))).((.....)).......))))).(((((.....)))))...... ( -19.10, z-score = -1.38, R) >droPer1.super_19 915979 102 + 1869541 --------------GUUUCCCUCGUUUCUGUGUGUUUUAAUUUAUGCAAAUAAAUUUCCACUUUGCACAGAAAUCAAAAUGAAUUAAGUGCGAGUGAUAGAAAAGUUGUCGUGUUG --------------......(((((((((((((((...(((((((....)))))))...))...)))))))).((.....)).......))))).(((((.....)))))...... ( -19.10, z-score = -1.38, R) >droVir3.scaffold_13047 8426152 89 + 19223366 ----------------------UUUUAUUUUGCAUUUUAAUUUAUGCAAAUAAAUUUCCACUUUGCACUGAAAUCAAAAUGAAUUAAGUGCGAGUGAUAGAAAUGUUGUUG----- ----------------------......(((((((........)))))))...((((((((((.(((((.((.((.....)).)).))))))))))...))))).......----- ( -19.80, z-score = -2.40, R) >droMoj3.scaffold_6540 30990791 94 - 34148556 --------------GUUUUCUACAUUU---UGCAUUUUAAUUUAUGCAAAUAAAUUUCCACUUUGCACAGAAAUCAAAAUGAAUUAAGUGCGAGUGAUAGAAAUGCUGUUG----- --------------((((((((...((---(((((........)))))))........(((((.((((..((.((.....)).))..))))))))).)))))).)).....----- ( -21.00, z-score = -2.03, R) >droGri2.scaffold_14624 3652331 91 + 4233967 -----------------UUUUUCGUUU---CGCAUUUUAAUUUAUGCAAAUAAAUUUCCACUUUGCACAGAAAUCAAAAUGAAUUAAGUGCGAGUGAUAGAUAUGUUGUUG----- -----------------.....(((.(---(((((........))))...........(((((.((((..((.((.....)).))..)))))))))...)).)))......----- ( -16.70, z-score = -0.80, R) >consensus _________________UACC_UUUUUUUCUGUAUUUUAAUUUAUGCAAAUAAAUUUCCACUUUGCACUGAAAUCAAAAUGAAUUAAGUGCGAGUGAUAGAAAAGUUGUCGUGUUG ......................(((((((.(((((........)))))..........(((((.((((..((.((.....)).))..)))))))))..)))))))........... (-13.63 = -13.68 + 0.05)
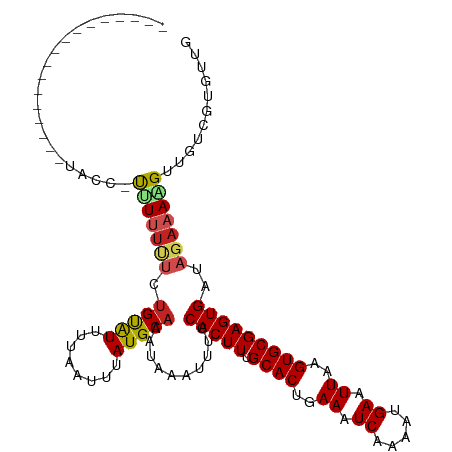
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:01:44 2011