| Sequence ID | dm3.chr2L |
|---|---|
| Location | 6,821,640 – 6,821,730 |
| Length | 90 |
| Max. P | 0.854063 |

| Location | 6,821,640 – 6,821,730 |
|---|---|
| Length | 90 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 73.17 |
| Shannon entropy | 0.49709 |
| G+C content | 0.47315 |
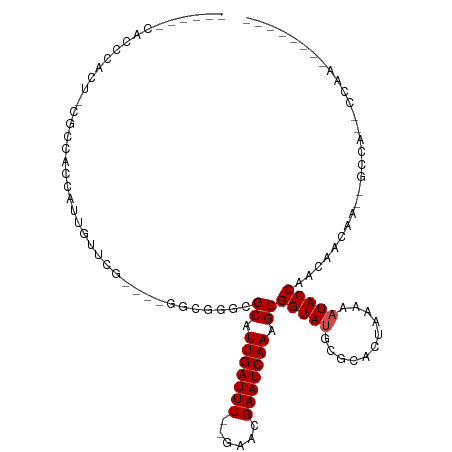
| Mean single sequence MFE | -24.97 |
| Consensus MFE | -11.57 |
| Energy contribution | -11.75 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.854063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
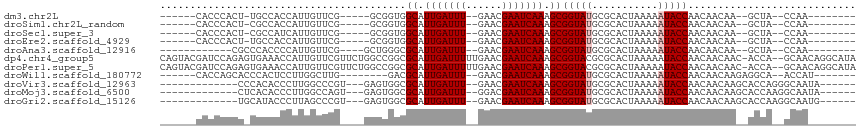
>dm3.chr2L 6821640 90 + 23011544 ------CACCCACU-UGCCACCAUUGUUCG-----GCGGUGGCAUUGAUUU--GAACGAAUCAAAGCGGUAUGCGCACUAAAAAUACCAACAACAA--GCUA--CCAA-------- ------........-.(((..........)-----))((((((.(((((((--....)))))))...(((((...........)))))........--))))--))..-------- ( -24.30, z-score = -2.07, R) >droSim1.chr2L_random 389812 90 + 909653 ------CACCCACU-CGCCACCAUUGUUCG-----GCGGUGGCAUUGAUUU--GAACGAAUCAAAGCGGUAUGCGCACUAAAAAUACCAACAACAA--GCUA--CCAA-------- ------........-.(((..........)-----))((((((.(((((((--....)))))))...(((((...........)))))........--))))--))..-------- ( -24.60, z-score = -2.31, R) >droSec1.super_3 2361545 90 + 7220098 ------CACCCACU-CGCCAUCAUUGUUCG-----GCGGUGGCAUUGAUUU--GAACGAAUCAAAGCGGUAUGCGCACUAAAAAUACCAACAACAA--GCUA--CCAA-------- ------........-.(((..........)-----))((((((.(((((((--....)))))))...(((((...........)))))........--))))--))..-------- ( -24.60, z-score = -2.39, R) >droEre2.scaffold_4929 15739510 90 + 26641161 ------CACCCACU-UGCCACCAUUGUUCG-----GCGGUGGCAUUGAUUU--GAACGAAUCAAAGCGGUAUGCGCACUAAAAAUACCAACAACAA--GCUA--CCAA-------- ------........-.(((..........)-----))((((((.(((((((--....)))))))...(((((...........)))))........--))))--))..-------- ( -24.30, z-score = -2.07, R) >droAna3.scaffold_12916 8407670 86 + 16180835 ------------CGCCCACCCCAUUGUUCG----GCUGGGCGCAUUGAUUU--GAACGAAUCAAAGCGGUAUGCGCACUAAAAAUACCAACAACAA--GCUA--CCAA-------- ------------.(((((.((........)----).)))))((.(((((((--....))))))).))((((.((......................--))))--))..-------- ( -25.55, z-score = -2.58, R) >dp4.chr4_group5 309515 113 + 2436548 CAGUACGAUCCAGAGUGAAACCAUUGUUCGUUCUGGCCGGCGCAUUGAUUUUUGAACGAAUCAAAGCGGUACGCGCACUAAAAAUACCAACAACAAC-ACCA--GCAACAGGCAUA (((.((((...(.((((....)))).))))).)))(((.((((.(((((((......))))))).))((((.............)))).........-....--))....)))... ( -25.42, z-score = -0.70, R) >droPer1.super_5 4806721 113 + 6813705 CAGUACGAUCCAGAGUGAAACCAUUGUUCGUUCUGGCCGGCGCAUUGAUUUUUGAACGAAUCAAAGCGGUACGCGCACUAAAAAUACCAACAACAAC-ACCA--GCAACAGGCAUA (((.((((...(.((((....)))).))))).)))(((.((((.(((((((......))))))).))((((.............)))).........-....--))....)))... ( -25.42, z-score = -0.70, R) >droWil1.scaffold_180772 1969509 91 - 8906247 ------CACCAGCACCCACUCCUUGGCUUG--------GACGCAUUGAUUU--GAACGAAUCAAAGCGGUAUGCGCACUAAAAAUACCAACAACAAGAGGCA--ACCAU------- ------.....((..(((.....)))((((--------...((.(((((((--....))))))).))(((((...........))))).....))))..)).--.....------- ( -20.00, z-score = -1.52, R) >droVir3.scaffold_12963 5340910 92 - 20206255 -------------CCCACACCCUUGGCCCGU---GAGUGGCGCAUUGAUUU--GAACGAAUCAAAGCGGUAUGCGCACUAAAAAUACCAACAACAAGCACCAGGGCAAUA------ -------------(((.....((((((((..---..).)))((.(((((((--....))))))).))(((((...........))))).....)))).....))).....------ ( -27.20, z-score = -2.00, R) >droMoj3.scaffold_6500 14533976 92 - 32352404 -------------CUCACACCCUUGGCCAGU---GAGUGGCGCAUUGAUUU--GGACGAAUCAAAGCGGUAUGCGCACUAAAAAUACCAACAACAAGCACCAAGGCAAUA------ -------------.......(((((((((..---...))))((.(((((((--....))))))).))(((((...........)))))............))))).....------ ( -24.10, z-score = -1.19, R) >droGri2.scaffold_15126 4504756 92 + 8399593 -------------UGCAUACCCUUAGCCCGU---GAGUGGCGCAUUGAUUU--GAACGAAUCAAAGCGGUAUGCGCACUAAAAAUACCAACAACAAGCACCAAGGCAAUG------ -------------.(((((((....((((..---..).)))((.(((((((--....))))))).)))))))))......................((......))....------ ( -29.20, z-score = -3.52, R) >consensus ______CACCCACU_CGCCACCAUUGUUCG____GGCGGGCGCAUUGAUUU__GAACGAAUCAAAGCGGUAUGCGCACUAAAAAUACCAACAACAA__GCCA__CCAA________ .........................................((.(((((((......))))))).))(((((...........)))))............................ (-11.57 = -11.75 + 0.18)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:22:13 2011