
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 658,537 – 658,639 |
| Length | 102 |
| Max. P | 0.994421 |

| Location | 658,537 – 658,639 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 77.19 |
| Shannon entropy | 0.43956 |
| G+C content | 0.57479 |
| Mean single sequence MFE | -38.26 |
| Consensus MFE | -27.87 |
| Energy contribution | -30.37 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.949445 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
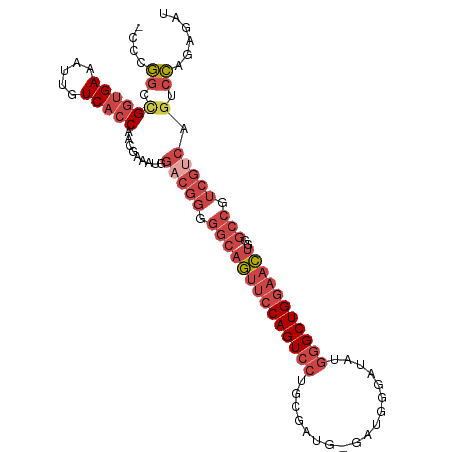
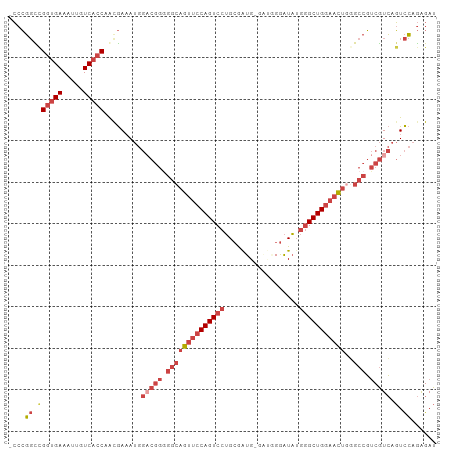
>dm3.chr2L 658537 102 + 23011544 -CCCGGCCGGUGAAAUUGUCACCAGCGAAAUGGACGGAGGCAGUUCCAGUCCUGCGAUGCGAUGGGAUAUGGGCUGGAACUGGGCCGUCGUCAGUCCAGAGAU -....((.(((((.....))))).))((....((((..((((((((((((((....((.(....).))..)))))))))))..)))..))))..))....... ( -41.70, z-score = -1.73, R) >droAna3.scaffold_12916 1649821 88 + 16180835 GGAGUCUCGUGGAAACUGUCGCCAACAACA-----------AAUGCCAGUCCAG-AGUGCGACAGAGUU---GCUGAAUUCACGCCCACAUGACUUAAAGGAU .(((((..((((...((((((((....((.-----------.......))....-.).)))))))....---((((....)).))))))..)))))....... ( -23.50, z-score = -0.89, R) >droEre2.scaffold_4929 717107 97 + 26641161 -CCCGGUUGGUGAAAUUGUCACCAAUGAAAUGGACGGGGGCAGUUCCAGUCCUGCGAUG-----GGAUAUGGGCUGGAACUUGGCCGUCGUCAGUCCAGAGAU -....((((((((.....))))))))......(((((.((((((((((((((.......-----......)))))))))))..))).)))))........... ( -41.92, z-score = -2.86, R) >droYak2.chr2L 649386 97 + 22324452 -CCCGGUCGGUGAAAUUGUCACCAAUGAAAUGGCCGGGGGCAGUUCCAGUCCUGCGGUG-----GGAUAUGGGCUGGAACUGGGCCGUCGUCAGUCCAGAGAU -((((((((((((.....)))))........)))))))((((((((((((((.......-----......))))))))))))(((....)))...))...... ( -40.72, z-score = -1.66, R) >droSec1.super_14 640399 102 + 2068291 -CCCGGCCGGUGAAAUUGUCACCAACGAAAUGGACGGGGGCAGUUCCAGUCCUGCGAUGGGGUGGGAUAUGGGCUGGAACUGGGCCGUCGUCAGUCCAGAGAU -...((.((((((.....))))).........(((((.((((((((((((((..................)))))))))))..))).))))).).))...... ( -40.77, z-score = -0.91, R) >droSim1.chr2L 669470 102 + 22036055 -CCCGGCCGGUGAAAUUGUCACCAACGAAAUGGACGGGGGCAGUUCCAGUCCUGCGAUGGGAUGGGAUAUGGGCUGGAACUGUGCCGUCGUCAGUCCAGAGAU -...((.((((((.....))))).........((((((.(((((((((((((..................))))))))))))).))..)))).).))...... ( -40.97, z-score = -1.44, R) >consensus _CCCGGCCGGUGAAAUUGUCACCAACGAAAUGGACGGGGGCAGUUCCAGUCCUGCGAUG_GAUGGGAUAUGGGCUGGAACUGGGCCGUCGUCAGUCCAGAGAU ....((.((((((.....))))).........(((((.((((((((((((((..................)))))))))))..))).))))).).))...... (-27.87 = -30.37 + 2.50)

| Location | 658,537 – 658,639 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 77.19 |
| Shannon entropy | 0.43956 |
| G+C content | 0.57479 |
| Mean single sequence MFE | -34.33 |
| Consensus MFE | -25.48 |
| Energy contribution | -26.95 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.70 |
| SVM RNA-class probability | 0.994421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
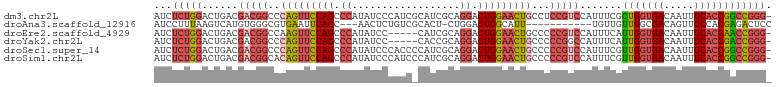
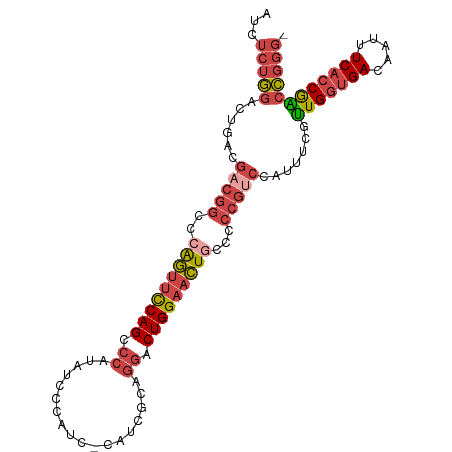
>dm3.chr2L 658537 102 - 23011544 AUCUCUGGACUGACGACGGCCCAGUUCCAGCCCAUAUCCCAUCGCAUCGCAGGACUGGAACUGCCUCCGUCCAUUUCGCUGGUGACAAUUUCACCGGCCGGG- .........(((..(((((..(((((((((.((..................)).)))))))))...)))))......((((((((.....))))))))))).- ( -38.27, z-score = -2.64, R) >droAna3.scaffold_12916 1649821 88 - 16180835 AUCCUUUAAGUCAUGUGGGCGUGAAUUCAGC---AACUCUGUCGCACU-CUGGACUGGCAUU-----------UGUUGUUGGCGACAGUUUCCACGAGACUCC ........((((.(((((((.((....))))---....(((((((...-...(((.......-----------.)))....)))))))...))))).)))).. ( -25.40, z-score = -0.75, R) >droEre2.scaffold_4929 717107 97 - 26641161 AUCUCUGGACUGACGACGGCCAAGUUCCAGCCCAUAUCC-----CAUCGCAGGACUGGAACUGCCCCCGUCCAUUUCAUUGGUGACAAUUUCACCAACCGGG- ....((((...((((..(((..((((((((.((......-----.......)).)))))))))))..)))).......(((((((.....))))))))))).- ( -35.12, z-score = -3.40, R) >droYak2.chr2L 649386 97 - 22324452 AUCUCUGGACUGACGACGGCCCAGUUCCAGCCCAUAUCC-----CACCGCAGGACUGGAACUGCCCCCGGCCAUUUCAUUGGUGACAAUUUCACCGACCGGG- ....((((.........(((((((((((((.((......-----.......)).))))))))).....))))......(((((((.....))))))))))).- ( -35.62, z-score = -2.89, R) >droSec1.super_14 640399 102 - 2068291 AUCUCUGGACUGACGACGGCCCAGUUCCAGCCCAUAUCCCACCCCAUCGCAGGACUGGAACUGCCCCCGUCCAUUUCGUUGGUGACAAUUUCACCGGCCGGG- .........(((..(((((..(((((((((.((..................)).)))))))))...)))))......((((((((.....))))))))))).- ( -35.77, z-score = -2.04, R) >droSim1.chr2L 669470 102 - 22036055 AUCUCUGGACUGACGACGGCACAGUUCCAGCCCAUAUCCCAUCCCAUCGCAGGACUGGAACUGCCCCCGUCCAUUUCGUUGGUGACAAUUUCACCGGCCGGG- .........(((..(((((..(((((((((.((..................)).)))))))))...)))))......((((((((.....))))))))))).- ( -35.77, z-score = -2.08, R) >consensus AUCUCUGGACUGACGACGGCCCAGUUCCAGCCCAUAUCCCAUC_CAUCGCAGGACUGGAACUGCCCCCGUCCAUUUCGUUGGUGACAAUUUCACCGACCGGG_ ....((((......(((((..(((((((((.((..................)).)))))))))...))))).......(((((((.....))))))))))).. (-25.48 = -26.95 + 1.48)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:06:33 2011