| Sequence ID | dm3.chr3R |
|---|---|
| Location | 5,699,838 – 5,699,913 |
| Length | 75 |
| Max. P | 0.995160 |

| Location | 5,699,838 – 5,699,913 |
|---|---|
| Length | 75 |
| Sequences | 7 |
| Columns | 83 |
| Reading direction | forward |
| Mean pairwise identity | 78.62 |
| Shannon entropy | 0.39492 |
| G+C content | 0.47827 |
| Mean single sequence MFE | -20.41 |
| Consensus MFE | -15.35 |
| Energy contribution | -15.51 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.06 |
| Mean z-score | -0.94 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.547617 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
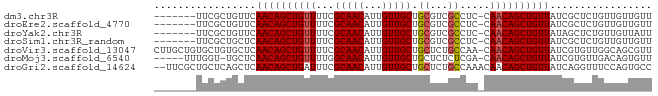
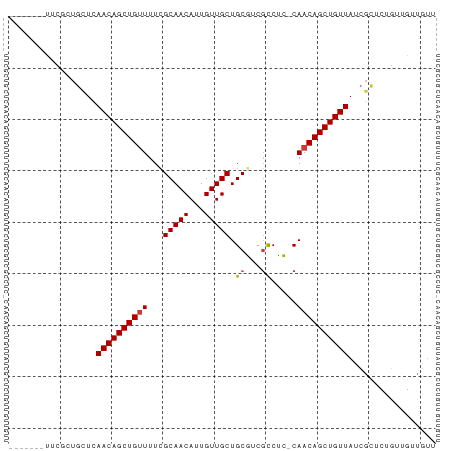
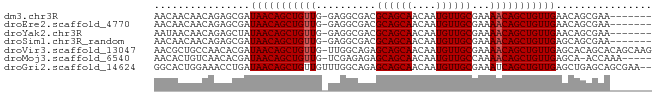
>dm3.chr3R 5699838 75 + 27905053 -------UUCGCUGUUCAACAGCUGUUUUCGCAACAUUGUUGCUGCGUCGCCUC-CAACAGCUGUUAUCGCUCUGUUGUUGUU -------...((.((..((((((((((..((((.((....)).)))).......-.)))))))))))).))............ ( -18.30, z-score = -1.08, R) >droEre2.scaffold_4770 1840370 75 - 17746568 -------UUCGCUGUUCAACAGCUGUUUUCGCAACAUUGUUGCUGCGUCGCCUC-CAACAGCUGUUAUCGCUCUGUUGUUGUU -------...((.((..((((((((((..((((.((....)).)))).......-.)))))))))))).))............ ( -18.30, z-score = -1.08, R) >droYak2.chr3R 9743139 75 + 28832112 -------UUCGCUGUUCAACAGCUGUUUUCGCAACAUUGUUGCUGCGUCGCCUC-CAACAGCUGUUAUAGCUCUGUUGUUAUU -------...(((((..((((((((((..((((.((....)).)))).......-.)))))))))))))))............ ( -22.50, z-score = -3.05, R) >droSim1.chr3R_random 370212 75 + 1307089 -------UUCGCUGCUCAACAGCUGUUUUCGCAACAUUGUUGCUGCGUCGCCUC-CAACAGCUGUUAUCGCUCUGUUGUUGUU -------......((..((((((((((..((((.((....)).)))).......-.))))))))))...))............ ( -18.50, z-score = -0.82, R) >droVir3.scaffold_13047 8375641 82 - 19223366 CUUGCUGUGCUGUGCUCAACAGCUGUUUUCGCAACAUUGUUGCUGCUCUGCCAA-CAACAGCUGUUAUCGUGUUGGCAGCGUU ...(((((...(..(..((((((((((...(((((...))))).((...))...-.))))))))))...)..)..)))))... ( -25.50, z-score = -0.20, R) >droMoj3.scaffold_6540 30944007 76 + 34148556 -----UUUGGU-UGCUCAACAGCUGUUUUGGCAACAUUGUUGCUGCUCUCUCGA-CAACAGCUGUUAUCGUGUUGACAGUGUU -----....((-((.....((((..(..((....))..)..))))......)))-)((((.((((((......)))))))))) ( -18.90, z-score = 0.12, R) >droGri2.scaffold_14624 3594131 81 - 4233967 --UUCGCUGCUCAGCUCAACAGCUGAUUUCGCAACAUUGUUGCUGCUCUGCCAAACAACAGCUGUUAUCAGGUUUCCAGUGCC --..(((((...((((.((((((((.....(((((...))))).((...)).......))))))))....))))..))))).. ( -20.90, z-score = -0.46, R) >consensus _______UUCGCUGCUCAACAGCUGUUUUCGCAACAUUGUUGCUGCGUCGCCUC_CAACAGCUGUUAUCGCUCUGUUGUUGUU .................((((((((((...(((((...))))).((...)).....))))))))))................. (-15.35 = -15.51 + 0.16)
| Location | 5,699,838 – 5,699,913 |
|---|---|
| Length | 75 |
| Sequences | 7 |
| Columns | 83 |
| Reading direction | reverse |
| Mean pairwise identity | 78.62 |
| Shannon entropy | 0.39492 |
| G+C content | 0.47827 |
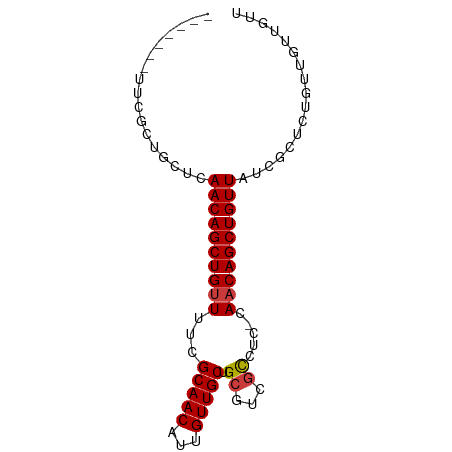
| Mean single sequence MFE | -25.50 |
| Consensus MFE | -19.35 |
| Energy contribution | -19.49 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.77 |
| SVM RNA-class probability | 0.995160 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 5699838 75 - 27905053 AACAACAACAGAGCGAUAACAGCUGUUG-GAGGCGACGCAGCAACAAUGUUGCGAAAACAGCUGUUGAACAGCGAA------- ............((..(((((((((((.-..(....)((((((....))))))...)))))))))))....))...------- ( -25.20, z-score = -3.25, R) >droEre2.scaffold_4770 1840370 75 + 17746568 AACAACAACAGAGCGAUAACAGCUGUUG-GAGGCGACGCAGCAACAAUGUUGCGAAAACAGCUGUUGAACAGCGAA------- ............((..(((((((((((.-..(....)((((((....))))))...)))))))))))....))...------- ( -25.20, z-score = -3.25, R) >droYak2.chr3R 9743139 75 - 28832112 AAUAACAACAGAGCUAUAACAGCUGUUG-GAGGCGACGCAGCAACAAUGUUGCGAAAACAGCUGUUGAACAGCGAA------- ............(((.(((((((((((.-..(....)((((((....))))))...)))))))))))...)))...------- ( -25.80, z-score = -3.76, R) >droSim1.chr3R_random 370212 75 - 1307089 AACAACAACAGAGCGAUAACAGCUGUUG-GAGGCGACGCAGCAACAAUGUUGCGAAAACAGCUGUUGAGCAGCGAA------- ............((..(((((((((((.-..(....)((((((....))))))...))))))))))).))......------- ( -26.00, z-score = -2.99, R) >droVir3.scaffold_13047 8375641 82 + 19223366 AACGCUGCCAACACGAUAACAGCUGUUG-UUGGCAGAGCAGCAACAAUGUUGCGAAAACAGCUGUUGAGCACAGCACAGCAAG ....(((((((((((........)).))-))))))).((.(((((...))))).......(((((.....)))))...))... ( -27.80, z-score = -1.49, R) >droMoj3.scaffold_6540 30944007 76 - 34148556 AACACUGUCAACACGAUAACAGCUGUUG-UCGAGAGAGCAGCAACAAUGUUGCCAAAACAGCUGUUGAGCA-ACCAAA----- ................(((((((((((.-((....))((((((....))))))...)))))))))))....-......----- ( -23.40, z-score = -2.51, R) >droGri2.scaffold_14624 3594131 81 + 4233967 GGCACUGGAAACCUGAUAACAGCUGUUGUUUGGCAGAGCAGCAACAAUGUUGCGAAAUCAGCUGUUGAGCUGAGCAGCGAA-- ......(....).........(((((((((..((((.((((((....))))))(....)..))))..)))..))))))...-- ( -25.10, z-score = -0.44, R) >consensus AACAACAACAGAGCGAUAACAGCUGUUG_GAGGCGACGCAGCAACAAUGUUGCGAAAACAGCUGUUGAGCAGCGAA_______ ................(((((((((((..........((((((....))))))...)))))))))))................ (-19.35 = -19.49 + 0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:01:37 2011