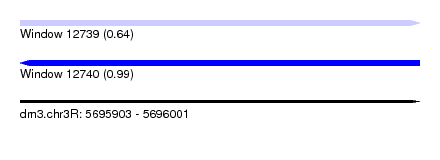
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 5,695,903 – 5,696,001 |
| Length | 98 |
| Max. P | 0.990170 |

| Location | 5,695,903 – 5,696,001 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 78.33 |
| Shannon entropy | 0.40551 |
| G+C content | 0.43743 |
| Mean single sequence MFE | -22.07 |
| Consensus MFE | -11.41 |
| Energy contribution | -14.37 |
| Covariance contribution | 2.96 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.641156 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
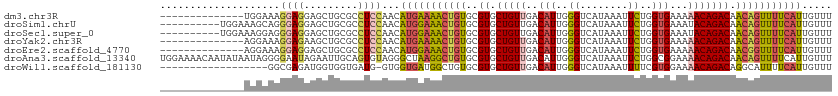
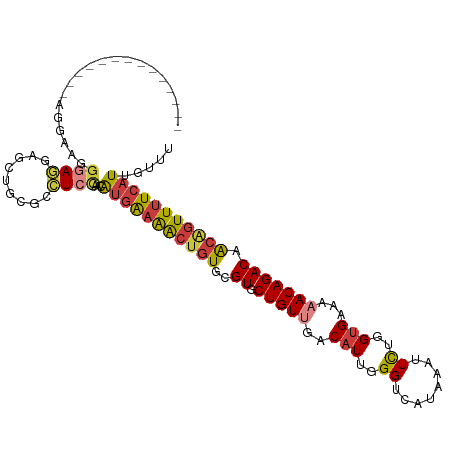
>dm3.chr3R 5695903 98 + 27905053 AAACAAUGAAAACUGUUGUCUGUUUUUCACCAGAAUUUAUGACCCAAUGUCAACAGCACGCACAGUUUUCAUGUUGGAGGCGCAGCUCCUCCUUUCCA-------------- .((((.(((((((((((((((((..(((....)))....((((.....)))))))).))).))))))))))))))(((((.......)))))......-------------- ( -29.10, z-score = -3.36, R) >droSim1.chrU 889334 102 + 15797150 AAACAAUGAAAACUGUUGUCUGUAUUUCACCAGAAUUUAUGACCCAAUGUCAACAGCACGCACAGUUUCCAUGUUGGAGGCGCAGCUCCUCCCUGCUUUCCA---------- .((((.((.((((((((((((((..(((....)))....((((.....)))))))).))).))))))).))))))(((((.((((.......))))))))).---------- ( -27.60, z-score = -2.49, R) >droSec1.super_0 4899215 102 - 21120651 AAACAAUGAAAACUGUUGUCUGUAUUUCACCAGAAUUUAUGACCCAAUGUCAACAGCACGCACAGUUUCCAUGUUGGAGGCGCAGCUCCUCCCUCCUUUCCA---------- .((((.((.((((((((((((((..(((....)))....((((.....)))))))).))).))))))).))))))(((((.(.((...))))))))......---------- ( -25.20, z-score = -2.27, R) >droYak2.chr3R 9738890 98 + 28832112 AAACAAUGAAAACUGUUGUCUGUUUUUCACCAGAAUUUAUGACCCAAUGUCAACAGCACGCACAGUUUUCAUGUUGGAGGCGCAGCUUCUCCUUUCCU-------------- .((((.(((((((((((((((((..(((....)))....((((.....)))))))).))).))))))))))))))(((((.......)))))......-------------- ( -27.00, z-score = -2.70, R) >droEre2.scaffold_4770 1836094 98 - 17746568 AAACAAUGAAAACCGUUGUCUGUUUUUCACCAGAAUUUAUGACCCAAUGUCAACAGCACGCACAGUUUCCAUGUUGGAGGCGCAGCUCCUCCUUUCCU-------------- .((((.((.((((.(((((((((..(((....)))....((((.....)))))))).))).)).)))).))))))(((((.......)))))......-------------- ( -20.10, z-score = -0.70, R) >droAna3.scaffold_13340 6255505 112 + 23697760 AAACAAUGAAAACUGUUGUCUGUUUUCCGCCAGAAUUUAUGACCCAAUGUCAACAGCACGCACAGCCUUAGCCCUACACUGCAAUUCUAUUCCCCUAUUAUAUUGUUUUCCA (((((((((((((........))))))....((((((..((((.....))))...(((......((....)).......))))))))).............))))))).... ( -15.92, z-score = -1.09, R) >droWil1.scaffold_181130 267210 93 - 16660200 AAACAAUGAAAAUGCCUGUCUGUUUUCCACGAAAAUUUAUGACCCAAUGUCAACAGCACGCACAGCCAUCACCAC-CAUCACCACCAUCUCGCC------------------ ............(((.((.((((((((...)))).....((((.....)))))))))).))).............-..................------------------ ( -9.60, z-score = -0.05, R) >consensus AAACAAUGAAAACUGUUGUCUGUUUUUCACCAGAAUUUAUGACCCAAUGUCAACAGCACGCACAGUUUUCAUGUUGGAGGCGCAGCUCCUCCCUUCCU______________ .....(((.((((((((((((((..(((....)))....((((.....)))))))).))).))))))).)))...(((((.......))))).................... (-11.41 = -14.37 + 2.96)

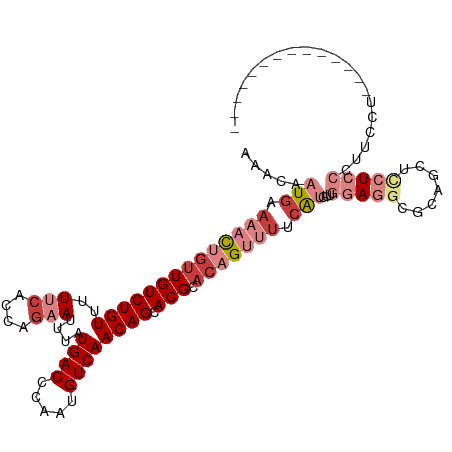
| Location | 5,695,903 – 5,696,001 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 78.33 |
| Shannon entropy | 0.40551 |
| G+C content | 0.43743 |
| Mean single sequence MFE | -30.87 |
| Consensus MFE | -23.94 |
| Energy contribution | -24.30 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.990170 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 5695903 98 - 27905053 --------------UGGAAAGGAGGAGCUGCGCCUCCAACAUGAAAACUGUGCGUGCUGUUGACAUUGGGUCAUAAAUUCUGGUGAAAAACAGACAACAGUUUUCAUUGUUU --------------......(((((.......)))))((((((((((((((..((.(((((..(((..((........))..)))...))))))).)))))))))).)))). ( -35.40, z-score = -3.80, R) >droSim1.chrU 889334 102 - 15797150 ----------UGGAAAGCAGGGAGGAGCUGCGCCUCCAACAUGGAAACUGUGCGUGCUGUUGACAUUGGGUCAUAAAUUCUGGUGAAAUACAGACAACAGUUUUCAUUGUUU ----------....(((((((((((.......)))))....((((((((((..((.((((...(((..((........))..)))....)))))).)))))))))))))))) ( -33.50, z-score = -2.26, R) >droSec1.super_0 4899215 102 + 21120651 ----------UGGAAAGGAGGGAGGAGCUGCGCCUCCAACAUGGAAACUGUGCGUGCUGUUGACAUUGGGUCAUAAAUUCUGGUGAAAUACAGACAACAGUUUUCAUUGUUU ----------..........(((((.......)))))((((((((((((((..((.((((...(((..((........))..)))....)))))).)))))))))).)))). ( -33.00, z-score = -2.30, R) >droYak2.chr3R 9738890 98 - 28832112 --------------AGGAAAGGAGAAGCUGCGCCUCCAACAUGAAAACUGUGCGUGCUGUUGACAUUGGGUCAUAAAUUCUGGUGAAAAACAGACAACAGUUUUCAUUGUUU --------------......((((.........))))((((((((((((((..((.(((((..(((..((........))..)))...))))))).)))))))))).)))). ( -31.80, z-score = -2.81, R) >droEre2.scaffold_4770 1836094 98 + 17746568 --------------AGGAAAGGAGGAGCUGCGCCUCCAACAUGGAAACUGUGCGUGCUGUUGACAUUGGGUCAUAAAUUCUGGUGAAAAACAGACAACGGUUUUCAUUGUUU --------------......(((((.......)))))((((((((((((((..((.(((((..(((..((........))..)))...))))))).)))))))))).)))). ( -34.20, z-score = -3.12, R) >droAna3.scaffold_13340 6255505 112 - 23697760 UGGAAAACAAUAUAAUAGGGGAAUAGAAUUGCAGUGUAGGGCUAAGGCUGUGCGUGCUGUUGACAUUGGGUCAUAAAUUCUGGCGGAAAACAGACAACAGUUUUCAUUGUUU ....(((((((............((((((((((.((((.(((....))).)))))))...((((.....))))..)))))))...((((((........))))))))))))) ( -27.70, z-score = -1.63, R) >droWil1.scaffold_181130 267210 93 + 16660200 ------------------GGCGAGAUGGUGGUGAUG-GUGGUGAUGGCUGUGCGUGCUGUUGACAUUGGGUCAUAAAUUUUCGUGGAAAACAGACAGGCAUUUUCAUUGUUU ------------------..................-..((..((((..((((.(((((((..(((..((........))..)))...))))).)).))))..))))..)). ( -20.50, z-score = 0.75, R) >consensus ______________AGGAAGGGAGGAGCUGCGCCUCCAACAUGAAAACUGUGCGUGCUGUUGACAUUGGGUCAUAAAUUCUGGUGAAAAACAGACAACAGUUUUCAUUGUUU ....................((((.........))))...(((((((((((..((.(((((..(((..((........))..)))...))))))).)))))))))))..... (-23.94 = -24.30 + 0.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:01:36 2011