| Sequence ID | dm3.chr2L |
|---|---|
| Location | 6,815,617 – 6,815,745 |
| Length | 128 |
| Max. P | 0.781629 |

| Location | 6,815,617 – 6,815,745 |
|---|---|
| Length | 128 |
| Sequences | 10 |
| Columns | 143 |
| Reading direction | reverse |
| Mean pairwise identity | 73.38 |
| Shannon entropy | 0.55887 |
| G+C content | 0.33333 |
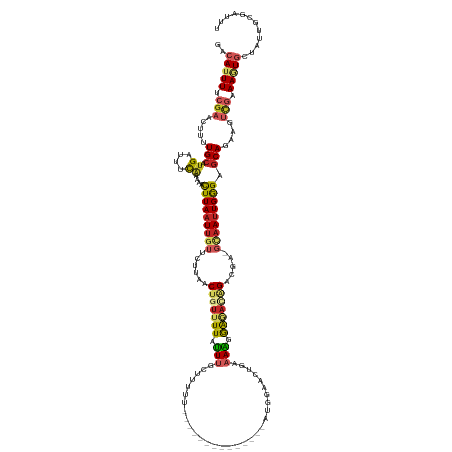
| Mean single sequence MFE | -32.09 |
| Consensus MFE | -10.51 |
| Energy contribution | -11.52 |
| Covariance contribution | 1.01 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.781629 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
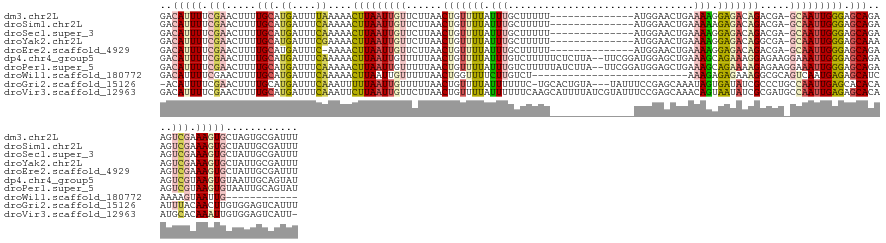
>dm3.chr2L 6815617 128 - 23011544 GACAUUUUCGAACUUUUGCAUGAUUUUAAAAACUUAAUUGUUCUUAACUGUUUUAUUUGCUUUUU--------------AUGGAACUGAAAAGGAGACAGACGA-GCAAUUGGGAGCAGAAGUCGAAAGUGCUAGUGCGAUUU ..(((((((((.(((((((.(........)..(((((((((((....(((((((.....((((((--------------(......))))))))))))))..))-))))))))).))))))))))))))))............ ( -36.30, z-score = -3.42, R) >droSim1.chr2L 6616618 128 - 22036055 GACAUUUUCGAACUUUUGCAUGAUUUCAAAAACUUAAUUGUUCUUAACUGUUUUAUUUGCUUUUU--------------AUGGAACUGAAAAAGAGACAGACGA-GCAAUUGGGAGCAGAAGUCGAAAGUGCUAUUGCGAUUU ..(((((((((.(((((((.((....))....(((((((((((....(((((((.(((.......--------------..........))).)))))))..))-))))))))).))))))))))))))))............ ( -36.03, z-score = -3.48, R) >droSec1.super_3 2354948 128 - 7220098 GACAUUUUCGAACUUUUGCAUGAUUUCAAAAACUUAAUUGUUCUUAACUGUUUUAUUUGCUUUUU--------------AUGGAACUGAAAAGGAGACAGACGA-GCAAUUGGGAGCAGAAGUCGAAAGUGCUAUUGCGAUUU ..(((((((((.(((((((.((....))....(((((((((((....(((((((.....((((((--------------(......))))))))))))))..))-))))))))).))))))))))))))))............ ( -39.10, z-score = -4.38, R) >droYak2.chr2L 16233134 128 + 22324452 GACAUUUUCGAACUUUUGCAUGAUUUCGAAAACUUAAUUGUUCUUAACUGUUUUAUUUGCUUUUU--------------AUGGAACUGAAAAGGAGACAGGCGA-GCAAUUGGGAGCAAAAGUCGAAAGUGCUAUUGCGAUUU ..(((((((((.(((((((.((....))....(((((((((((....(((((((.....((((((--------------(......))))))))))))))..))-))))))))).))))))))))))))))............ ( -39.40, z-score = -4.21, R) >droEre2.scaffold_4929 15732402 127 - 26641161 GACAUUUUCGAACUUUUGCAUGAUUUC-AAAACUUAAUUGUUCUUAACUGUUUUAUUUGCUUUUU--------------AUGGAACUGAAAAGGAGACAGACGA-GCAAUUGGGAGCAGAAGUCGAAAGUGCUAUUGCGAUUU ..(((((((((.(((((((.((....)-)...(((((((((((....(((((((.....((((((--------------(......))))))))))))))..))-))))))))).))))))))))))))))............ ( -39.10, z-score = -4.37, R) >dp4.chr4_group5 301360 141 - 2436548 GACAUUUUCGAACUUUUGCAUGAUUUCAAAAACUUAAUUGUUUUUAACUGUUUUAUUUGUCUUUUUCUCUUA--UUCGGAUGGAGCUGAAAGCAGAAAGGAGAAGGAAAUUGGGAGCAGAAGUCGUAAGUGUAAUUGCAGUAU .((((((.(((.(((((((.(((((((((((((......))))))..........(((.(((((((..(((.--.((((......)))))))..))))))).))))))))))...)))))))))).))))))........... ( -34.50, z-score = -1.40, R) >droPer1.super_5 4798511 141 - 6813705 GACAUUUUCGAACUUUUGCAUGAUUUCAAAAACUUAAUUGUUUUUAACUGUUUUAUUUGUCUUUUUAUCUUA--UUCGGAUGGAGCUGAAAGCAGAAAAGAGAAGGAAAUUGGGAGCAGAAGUCGUAAGUGUAAUUGCAGUAU .((((((.(((.(((((((.(((((((((((((......))))))..(((((((....((...(((((((..--...)))))))))..)))))))..........)))))))...)))))))))).))))))........... ( -34.30, z-score = -1.78, R) >droWil1.scaffold_180772 1957844 105 + 8906247 GACAUUUUCGAACUUUUGCAUGAUUUCAAAAACUUAAUUGUUUUUAACUGGUUUUCUUGUCU--------------------------AAAGAGAGAAAGGCGCAGUCAAUGAGAGCAUCAAAAGUAAUUG------------ ...........(((((((.(((.((((((((((......)))))).(((((((((..(.((.--------------------------...)).)..))))).))))....)))).)))))))))).....------------ ( -22.90, z-score = -1.02, R) >droGri2.scaffold_15126 4496877 138 - 8399593 -ACAUUUUCGAACUUUUGCAUGAUUUCAAAUUUUUAAUUGUUUUUAACUGUUUUAUUUUUUC-UGCACUGUA---UAUUUCCGAGCAAAUAGUGAUAUCGCCCUGCCAAUUGAGCACACAAUUUACAACUUGUGGAGUCAUUU -..................(((((((((.......((((((.....................-..((((((.---.............)))))).....((.(........).))..)))))).........))))))))).. ( -16.53, z-score = 1.45, R) >droVir3.scaffold_12963 5331720 142 + 20206255 GACAUUUUCGAACUUUUGCAUGAUUUCAAAUUCUUAAUUGUUCUUAACUGUUUUAUUUUUUCAAGCAUUUUAUCGUAUUUCCGAGCAAACAGUAAUAUCGCGAUGCCAAUUGAGAGCACAAUGCACAAAUUGUGGAGUCAUU- ...................((((((((...((((((((((........(((((.(.....).))))).......(((((..(((.............))).))))))))))))))).(((((......))))))))))))).- ( -22.72, z-score = 0.79, R) >consensus GACAUUUUCGAACUUUUGCAUGAUUUCAAAAACUUAAUUGUUCUUAACUGUUUUAUUUGCUUUUU______________AUGGAACUGAAAAGGAGACAGACGA_GCAAUUGGGAGCAGAAGUCGAAAGUGCUAUUGCGAUUU ..(((((.(((.....(((.((....))....(((((((((......(((((((.(((...............................))).))))))).....))))))))).)))....))).)))))............ (-10.51 = -11.52 + 1.01)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:22:12 2011