| Sequence ID | dm3.chr3R |
|---|---|
| Location | 5,676,693 – 5,676,808 |
| Length | 115 |
| Max. P | 0.726942 |

| Location | 5,676,693 – 5,676,808 |
|---|---|
| Length | 115 |
| Sequences | 12 |
| Columns | 133 |
| Reading direction | forward |
| Mean pairwise identity | 80.56 |
| Shannon entropy | 0.40520 |
| G+C content | 0.40073 |
| Mean single sequence MFE | -30.04 |
| Consensus MFE | -20.49 |
| Energy contribution | -20.34 |
| Covariance contribution | -0.15 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.586522 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
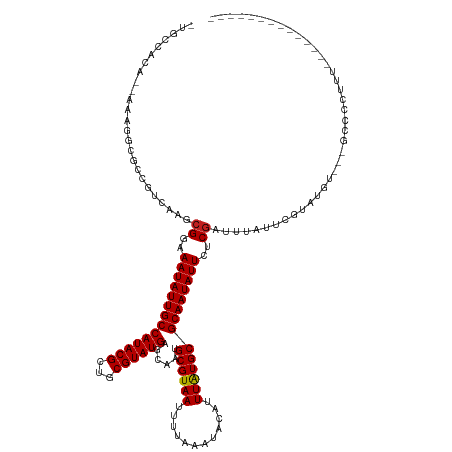
>dm3.chr3R 5676693 115 + 27905053 -------------AAAAGUGGC---ACAUACGAAUAAAUCGAGAAUAUUGCGCAUAAAUGUAUUUAAAAUUACGCAUUGCUCAUACGCAGCGUAUGGCAAUAUUUCCGCUUGACGGCGCCUUU--UGUGGCAG -------------..((((((.---.....(((.....))).((((((((((((....((((........))))...))).((((((...)))))))))))))))))))))......(((...--...))).. ( -30.90, z-score = -1.02, R) >droSec1.super_0 4880168 114 - 21120651 --------------AAAGGGGC---ACAUACGAAUAAAUUGAGAAUAUUGCGCAUAAAUGUAUUUAAAAUUACGCAUUGCUCAUACGCUGCGUAUGGCAAUAUUUCCGCUUGACGGCGCCUUU--UGUGGCAG --------------.(((((((---...............(..(((((((((((....((((........))))...))).((((((...))))))))))))))..)(((....)))))))))--)....... ( -30.10, z-score = -0.75, R) >droEre2.scaffold_4770 1804877 114 - 17746568 --------------AAAAGGGC---ACAUACGAAUAAAUCGAGAAUAUUGCGCAUAAAUGUAUUUAAAAUUACGCAUUGCUCAUACGCAGCGUAUGGCAAUAUUUCCGCUUGACGGCGCCUUU--UGUGGCAG --------------.(((((((---...............(..(((((((((((....((((........))))...))).((((((...))))))))))))))..)(((....)))))))))--)....... ( -30.80, z-score = -1.21, R) >droYak2.chr3R 9711750 114 + 28832112 --------------AAAGGGGC---ACAUACGAAUAAAUCGAGAAUAUUGCGCAUAAAUGUAUUUAAAAUUACGCAUUGCUCAUACGCAGCGUAUGGCAAUAUUUCCGCUUGACGGCGCCUUU--UGUGGCAG --------------.(((((((---...............(..(((((((((((....((((........))))...))).((((((...))))))))))))))..)(((....)))))))))--)....... ( -30.40, z-score = -0.90, R) >droSim1.chr3R 11835705 114 - 27517382 --------------AAAGGGGC---ACAUACGAAUAAAUCGAGAAUAUUGCGCAUAAAUGUAUUUAAAAUUACGCAUUGCUCAUACGCAGCGUAUGGCAAUAUUUCCGCUUGACGGCGCCUUU--UGUGGCAG --------------.(((((((---...............(..(((((((((((....((((........))))...))).((((((...))))))))))))))..)(((....)))))))))--)....... ( -30.40, z-score = -0.90, R) >droAna3.scaffold_13340 6225832 108 + 23697760 --------------AAAGGGCC---AC-UGCGAAUAAAUCGAGAAUAUUGCGCAUAAAUGUAUUUAAAAUUACGCAUUGCUCAUACGCAGCGUAUGGCAAUAUUUCCGCUUGACAGCGUCUUU--UGU----- --------------(((((((.---.(-(((((......((..(((((((((((....((((........))))...))).((((((...))))))))))))))..)).))).))).))))))--)..----- ( -27.70, z-score = -0.90, R) >dp4.chr2 18373282 98 - 30794189 -----------------------------AUAAAUAAAUCGAGAAUAUUGCGCAUAAAUGUAUUUAAAAUUACGCAUUGCUCAUACGCAACGUAUGGCAAUAUUUCCGCUCGACGGCGUCUUU--UGUG---- -----------------------------.........((((((((((((((((....((((........))))...))).((((((...))))))))))))))....)))))..........--....---- ( -22.20, z-score = -0.97, R) >droPer1.super_0 8053234 127 + 11822988 AAUGAGGGGCCGAAAGGGUGGCUGGAGAGACAAAUAAAUCGAGAAUAUUGCGCAUAAAUGUAUUUAAAAUUACGCAUUGCUCAUACGCAACGUAUGGCAAUAUUUCCGCUCGACGGCGCCUUU--UGUG---- ...((((.((((...((((((((.((............)).))(((((((((((....((((........))))...))).((((((...)))))))))))))).))))))..)))).)))).--....---- ( -40.70, z-score = -2.76, R) >droWil1.scaffold_181130 205486 117 - 16660200 --AAAAGAAACGGACAUCAUUCGGAAUCGGCAAAUAAAUCGAGAAUAUUGCGCAUAAAUGUAUUUAAAAUUACGCAUUGCUCAUACGCAGCGUAUGGCAAUAUUUCCGCUUCAUAGGGU-------------- --....(((.((((............((((........)))).(((((((((((....((((........))))...))).((((((...)))))))))))))))))).))).......-------------- ( -25.90, z-score = -1.09, R) >droVir3.scaffold_13047 8344764 114 - 19223366 -------------AAAGGGAGC---GCAAUGAUAUAAAUCGACAAUAUUGCGCAUAAAUGUGUUUAAAAUUACGCAUUGCUCAUACGCAGCGUAUGGCAAUAUUUCCGCUUCAUGGCACAUUA--UAUGCAU- -------------....(((((---(....(((....)))((.(((((((((((...((((((........))))))))).((((((...))))))))))))))))))))))...(((.....--..)))..- ( -30.60, z-score = -1.34, R) >droMoj3.scaffold_6540 30902553 114 + 34148556 -------------AAAAGCAGC---GCGACCAUAUAAAUCGACAAUAUUGCGCAUAAAUGUGUUUAAAAUUACGCAUUGCUCAUACGCAGCGUAUGGCAAUAUUUCCGCUUCAUGGCAUUUUA--UAUGCAU- -------------.........---.....((((((((..((.(((((((((((...((((((........))))))))).((((((...)))))))))))))))).(((....)))..))))--))))...- ( -28.40, z-score = -0.76, R) >droGri2.scaffold_14624 3551702 116 - 4233967 -------------AAAAGGAGC---GCGAUGAAGUAAAUCGACAAUAUUGCGCACAAAUGUGUUUAAAAUUACGCAUUGCUCAUACGCAGCGUAUGGCAAUAUUUCCGCUUGAUCGCAUUUUUGUUAUGCAG- -------------...(((((.---(((((.((((.....((.(((((((((((...((((((........))))))))).((((((...)))))))))))))))).)))).))))).))))).........- ( -32.40, z-score = -1.28, R) >consensus ______________AAAGGGGC___ACAUACAAAUAAAUCGAGAAUAUUGCGCAUAAAUGUAUUUAAAAUUACGCAUUGCUCAUACGCAGCGUAUGGCAAUAUUUCCGCUUGACGGCGCCUUU__UGUGGCA_ ........................................(..(((((((((((....((((........))))...))).((((((...))))))))))))))..)(((....)))................ (-20.49 = -20.34 + -0.15)



| Location | 5,676,693 – 5,676,808 |
|---|---|
| Length | 115 |
| Sequences | 12 |
| Columns | 133 |
| Reading direction | reverse |
| Mean pairwise identity | 80.56 |
| Shannon entropy | 0.40520 |
| G+C content | 0.40073 |
| Mean single sequence MFE | -29.02 |
| Consensus MFE | -20.49 |
| Energy contribution | -20.50 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.726942 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 5676693 115 - 27905053 CUGCCACA--AAAGGCGCCGUCAAGCGGAAAUAUUGCCAUACGCUGCGUAUGAGCAAUGCGUAAUUUUAAAUACAUUUAUGCGCAAUAUUCUCGAUUUAUUCGUAUGU---GCCACUUUU------------- ........--...(((((((.....((..((((((((((((((...))))))......((((((............))))))))))))))..)).......))...))---)))......------------- ( -29.90, z-score = -1.08, R) >droSec1.super_0 4880168 114 + 21120651 CUGCCACA--AAAGGCGCCGUCAAGCGGAAAUAUUGCCAUACGCAGCGUAUGAGCAAUGCGUAAUUUUAAAUACAUUUAUGCGCAAUAUUCUCAAUUUAUUCGUAUGU---GCCCCUUU-------------- ........--(((((.((...((.(((((...(((((((((((...)))))).)))))((((((............)))))).................))))).)).---)).)))))-------------- ( -27.30, z-score = -0.76, R) >droEre2.scaffold_4770 1804877 114 + 17746568 CUGCCACA--AAAGGCGCCGUCAAGCGGAAAUAUUGCCAUACGCUGCGUAUGAGCAAUGCGUAAUUUUAAAUACAUUUAUGCGCAAUAUUCUCGAUUUAUUCGUAUGU---GCCCUUUU-------------- ........--...(((((((.....((..((((((((((((((...))))))......((((((............))))))))))))))..)).......))...))---))).....-------------- ( -29.00, z-score = -0.91, R) >droYak2.chr3R 9711750 114 - 28832112 CUGCCACA--AAAGGCGCCGUCAAGCGGAAAUAUUGCCAUACGCUGCGUAUGAGCAAUGCGUAAUUUUAAAUACAUUUAUGCGCAAUAUUCUCGAUUUAUUCGUAUGU---GCCCCUUU-------------- ........--...(((((((.....((..((((((((((((((...))))))......((((((............))))))))))))))..)).......))...))---))).....-------------- ( -29.00, z-score = -0.99, R) >droSim1.chr3R 11835705 114 + 27517382 CUGCCACA--AAAGGCGCCGUCAAGCGGAAAUAUUGCCAUACGCUGCGUAUGAGCAAUGCGUAAUUUUAAAUACAUUUAUGCGCAAUAUUCUCGAUUUAUUCGUAUGU---GCCCCUUU-------------- ........--...(((((((.....((..((((((((((((((...))))))......((((((............))))))))))))))..)).......))...))---))).....-------------- ( -29.00, z-score = -0.99, R) >droAna3.scaffold_13340 6225832 108 - 23697760 -----ACA--AAAGACGCUGUCAAGCGGAAAUAUUGCCAUACGCUGCGUAUGAGCAAUGCGUAAUUUUAAAUACAUUUAUGCGCAAUAUUCUCGAUUUAUUCGCA-GU---GGCCCUUU-------------- -----...--((((.((((((.((.((..((((((((((((((...))))))......((((((............))))))))))))))..)).....)).)))-))---)...))))-------------- ( -29.50, z-score = -1.75, R) >dp4.chr2 18373282 98 + 30794189 ----CACA--AAAGACGCCGUCGAGCGGAAAUAUUGCCAUACGUUGCGUAUGAGCAAUGCGUAAUUUUAAAUACAUUUAUGCGCAAUAUUCUCGAUUUAUUUAU----------------------------- ----....--.........((((((....((((((((((((((((((......)))))).(((........)))...)))).))))))))))))))........----------------------------- ( -24.00, z-score = -1.47, R) >droPer1.super_0 8053234 127 - 11822988 ----CACA--AAAGGCGCCGUCGAGCGGAAAUAUUGCCAUACGUUGCGUAUGAGCAAUGCGUAAUUUUAAAUACAUUUAUGCGCAAUAUUCUCGAUUUAUUUGUCUCUCCAGCCACCCUUUCGGCCCCUCAUU ----....--..(((.((((..(((.((.((((((((((((((((((......)))))).(((........)))...)))).))))))))...(((......)))..........))))).)))).))).... ( -32.70, z-score = -2.09, R) >droWil1.scaffold_181130 205486 117 + 16660200 --------------ACCCUAUGAAGCGGAAAUAUUGCCAUACGCUGCGUAUGAGCAAUGCGUAAUUUUAAAUACAUUUAUGCGCAAUAUUCUCGAUUUAUUUGCCGAUUCCGAAUGAUGUCCGUUUCUUUU-- --------------.......((((((((((((((((((((((...))))))......((((((............))))))))))))))......(((((((.......)))))))..))))))))....-- ( -30.20, z-score = -2.61, R) >droVir3.scaffold_13047 8344764 114 + 19223366 -AUGCAUA--UAAUGUGCCAUGAAGCGGAAAUAUUGCCAUACGCUGCGUAUGAGCAAUGCGUAAUUUUAAACACAUUUAUGCGCAAUAUUGUCGAUUUAUAUCAUUGC---GCUCCCUUU------------- -..((((.--....))))...((((.(((..((((((((((((...)))))).)))))).....................(((((((..(((......)))..)))))---)))))))))------------- ( -29.10, z-score = -0.90, R) >droMoj3.scaffold_6540 30902553 114 - 34148556 -AUGCAUA--UAAAAUGCCAUGAAGCGGAAAUAUUGCCAUACGCUGCGUAUGAGCAAUGCGUAAUUUUAAACACAUUUAUGCGCAAUAUUGUCGAUUUAUAUGGUCGC---GCUGCUUUU------------- -..((((.--....))))...(((((((.((((((((((((((...))))))......((((((............))))))))))))))(.(((((.....))))))---.))))))).------------- ( -30.30, z-score = -0.47, R) >droGri2.scaffold_14624 3551702 116 + 4233967 -CUGCAUAACAAAAAUGCGAUCAAGCGGAAAUAUUGCCAUACGCUGCGUAUGAGCAAUGCGUAAUUUUAAACACAUUUGUGCGCAAUAUUGUCGAUUUACUUCAUCGC---GCUCCUUUU------------- -...............(((((.(((((..((((((((..((((((((......)))..)))))........(((....))).))))))))..)).....))).)))))---.........------------- ( -28.20, z-score = -0.73, R) >consensus _UGCCACA__AAAGGCGCCGUCAAGCGGAAAUAUUGCCAUACGCUGCGUAUGAGCAAUGCGUAAUUUUAAAUACAUUUAUGCGCAAUAUUCUCGAUUUAUUCGUAUGU___GCCCCUUU______________ .........................((..((((((((((((((...))))))......((((((............))))))))))))))..))....................................... (-20.49 = -20.50 + 0.01)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:01:32 2011