
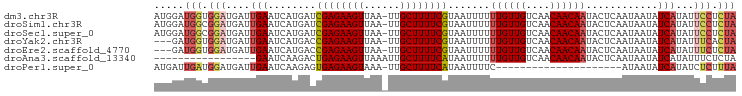
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 5,653,034 – 5,653,162 |
| Length | 128 |
| Max. P | 0.786645 |

| Location | 5,653,034 – 5,653,130 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 82.02 |
| Shannon entropy | 0.35769 |
| G+C content | 0.29037 |
| Mean single sequence MFE | -19.10 |
| Consensus MFE | -13.26 |
| Energy contribution | -13.93 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.786645 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 5653034 96 + 27905053 AUGGAUGGUGGAUGAUUGAAUCAUGAUCGAGAAGUUAA-UUGCUUUUCGUAAUUUUUUGUUGUCAACAACAAUACUCAAUAAUAUCAUAUUCCUCUA ..((((((((....(((((........((((((((...-..)))))))).......((((((....))))))...)))))..)))))...))).... ( -21.70, z-score = -1.64, R) >droSim1.chr3R 11824661 96 - 27517382 AUGGAUGGCGGAUGAUUGAAUCAUGAUCGAGAAGUUAA-UUGCUUUUCGUAAUUUUUUGUUGUCAACAACAAUACUCAAUAAUAUCAUAUUCCUCUA ..(((.....(((((((((........((((((((...-..)))))))).......((((((....))))))...)))))..))))....))).... ( -21.00, z-score = -1.52, R) >droSec1.super_0 4869166 96 - 21120651 AUGGAUGGCGGAUGAUUGAAUCAUGAUCGAGAAGUUAA-UUGCUUUUCGUAAUUUUUUGUUGUCAACAACAAUACUCAAUAAUAUCAUAUUCCUCUA ..(((.....(((((((((........((((((((...-..)))))))).......((((((....))))))...)))))..))))....))).... ( -21.00, z-score = -1.52, R) >droYak2.chr3R 9700375 93 + 28832112 ---GAUGGUGGAUGAUUGAAUCAUGACCGAGAAGUUAA-UUGCUUUUCGUAAUUUUUUGUUGUCAACAACAAUACUCAAUAAUAUCAUAUUUCACUA ---.((((((....(((((........((((((((...-..)))))))).......((((((....))))))...)))))..))))))......... ( -20.90, z-score = -1.97, R) >droEre2.scaffold_4770 1794455 93 - 17746568 ---GAUGGUGGAUGAUUGAAUCAUGACCGAGAAGUUAA-UUGCUUUUCGUAAUUUUUUGUUGUCAACAACAAUACUCAAUAAUAUCAUAUUUCUCUA ---.((((((....(((((........((((((((...-..)))))))).......((((((....))))))...)))))..))))))......... ( -20.90, z-score = -2.02, R) >droAna3.scaffold_13340 6215268 80 + 23697760 -----------------GAAUCAAGACUGAGAAGUUAAAUUGCUUUUCAUAAUUUUUUGUUGUCAACAACAAUACUCAAUAAUAUCAUAUUUCUCUA -----------------......(((.((((((((......)))))))).......((((((....)))))).....................))). ( -12.80, z-score = -1.76, R) >droPer1.super_0 8041427 75 + 11822988 AUGAUUGAUGGAUGAUUGAAUCAAGAGUGAGAAGUAAA-UUGCUUUUCAUAAUUUUC---------------------AUAAUAUCAUAUCUCUUUA (((((..(((((((((...))))...(((((((((...-..)))))))))....)))---------------------))...)))))......... ( -15.40, z-score = -0.87, R) >consensus AUGGAUGGUGGAUGAUUGAAUCAUGACCGAGAAGUUAA_UUGCUUUUCGUAAUUUUUUGUUGUCAACAACAAUACUCAAUAAUAUCAUAUUUCUCUA .....(((.(((....(((........((((((((......)))))))).......((((((....))))))............)))...))).))) (-13.26 = -13.93 + 0.67)

| Location | 5,653,069 – 5,653,162 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 96.88 |
| Shannon entropy | 0.05383 |
| G+C content | 0.26295 |
| Mean single sequence MFE | -8.16 |
| Consensus MFE | -7.70 |
| Energy contribution | -7.70 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.657450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 5653069 93 + 27905053 UAAUUGCUUUUCGUAAUUUUUUGUUGUCAACAACAAUACUCAAUAAUAUCAUAUUCCUCUAUAUAUCAUUCAAAUGAUCACAGUCAAAAAACG .((((((.....))))))..((((((....))))))............................(((((....)))))............... ( -8.00, z-score = -1.30, R) >droSim1.chr3R 11824696 93 - 27517382 UAAUUGCUUUUCGUAAUUUUUUGUUGUCAACAACAAUACUCAAUAAUAUCAUAUUCCUCUAUAUAUCAUUCAAAUGAUCACAGUCAAAAAACG .((((((.....))))))..((((((....))))))............................(((((....)))))............... ( -8.00, z-score = -1.30, R) >droSec1.super_0 4869201 93 - 21120651 UAAUUGCUUUUCGUAAUUUUUUGUUGUCAACAACAAUACUCAAUAAUAUCAUAUUCCUCUAUAUAUCAUUCAAAUGAUCACAGUCAAAAAACG .((((((.....))))))..((((((....))))))............................(((((....)))))............... ( -8.00, z-score = -1.30, R) >droYak2.chr3R 9700407 93 + 28832112 UAAUUGCUUUUCGUAAUUUUUUGUUGUCAACAACAAUACUCAAUAAUAUCAUAUUUCACUACAUCUCAUUCAAAUGAUCACAGUCAUAAAACG .((((((.....))))))..((((((....)))))).....................................(((((....)))))...... ( -8.90, z-score = -1.55, R) >droEre2.scaffold_4770 1794487 92 - 17746568 UAAUUGCUUUUCGUAAUUUUUUGUUGUCAACAACAAUACUCAAUAAUAUCAUAUUUCUCUACAUCUCAUUCAAAUGAUCACAGUCA-AAAACG .((((((.....))))))..((((((....))))))......................................((((....))))-...... ( -7.90, z-score = -1.36, R) >consensus UAAUUGCUUUUCGUAAUUUUUUGUUGUCAACAACAAUACUCAAUAAUAUCAUAUUCCUCUAUAUAUCAUUCAAAUGAUCACAGUCAAAAAACG .((((((.....))))))..((((((....))))))......................................((((....))))....... ( -7.70 = -7.70 + 0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:01:28 2011