| Sequence ID | dm3.chr3R |
|---|---|
| Location | 5,604,300 – 5,604,392 |
| Length | 92 |
| Max. P | 0.950605 |

| Location | 5,604,300 – 5,604,392 |
|---|---|
| Length | 92 |
| Sequences | 15 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 64.40 |
| Shannon entropy | 0.76264 |
| G+C content | 0.43886 |
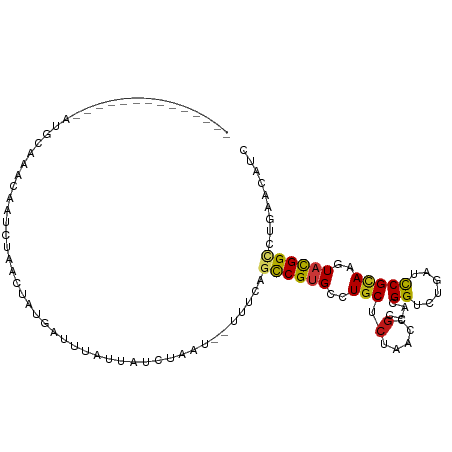
| Mean single sequence MFE | -20.92 |
| Consensus MFE | -7.49 |
| Energy contribution | -7.27 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.950605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 5604300 92 + 27905053 --------------AUGGAACCGAUCUAACUAUGAUAUACCAUCUCAU--UUUCAGCCGUGCCUGCUCUAACCGCCACGGUCUGAUCCGCAAGUAUGGCCUUAACAUC --------------((((.....(((.......)))...)))).....--.....(((((((.(((((..((((...))))..))...))).)))))))......... ( -21.40, z-score = -1.90, R) >droEre2.scaffold_4770 1745489 91 - 17746568 --------------AUGAAACCGAUGUAACUAUGAUAUACCAUCUUAU--UUU-AGCCGUGCCUGCUCUAACCGCCACGGUCUGAUCCGCAAGUACGGCCUGAACAUC --------------........(((((......(((.....)))....--.((-((((((((.(((((..((((...))))..))...))).)))))).))))))))) ( -23.90, z-score = -3.05, R) >droYak2.chr3R 9650514 91 + 28832112 --------------AUGGAACCGAUGUAAUCAUAAUAUACCAUCUCAU--UUU-AGCCGUGCCUGCUCUAACCGCCACGGUCUGAUCCGCAAGUACGGCCUGAACAUU --------------((((.....(((....)))......)))).....--(((-((((((((.(((((..((((...))))..))...))).)))))).))))).... ( -23.60, z-score = -2.28, R) >droSec1.super_0 4819364 92 - 21120651 --------------AUGGAACCGAACUAAAUAUGAUAUACCGUCUCAU--UUUCAGCCGUGCCUGCUCUAACCGCCACGGUCUGAUCCGCAAGUAUGGCCUUAACAUC --------------.(((((..((((...............)).))..--)))))(((((((.(((((..((((...))))..))...))).)))))))......... ( -19.06, z-score = -0.82, R) >droSim1.chr3R 11787788 92 - 27517382 --------------AUGGAACCGAACUAAAUAUGAUAUACCGUCUCAU--UUUCAGCCGUGCCUGCUCUAACCGCCACGGUCUGAUCCGCAAGUAUGGCCUUAACAUC --------------.(((((..((((...............)).))..--)))))(((((((.(((((..((((...))))..))...))).)))))))......... ( -19.06, z-score = -0.82, R) >dp4.chr2 18311906 89 - 30794189 -------------CAAGCAUAGAAUCUACUUAUGACUUGUU----UAA--UUCCAGCCGUGCCUGCUCUAACCGUCACGGUCUCAUCCGCAAGUACGGUCUGAACAUC -------------(((((((((.......))))).))))..----...--...(((((((((.(((....((((...)))).......))).)))))).)))...... ( -23.40, z-score = -2.84, R) >droPer1.super_0 7985980 89 + 11822988 -------------CAAGCAUAGAAUCUACUUAUGACUUGUU----UAA--UUCCAGCCGUGCCUGCUCUAACCGUCACGGUCUCAUCCGCAAGUACGGUCUGAACAUC -------------(((((((((.......))))).))))..----...--...(((((((((.(((....((((...)))).......))).)))))).)))...... ( -23.40, z-score = -2.84, R) >droAna3.scaffold_13340 6168739 88 + 23697760 ------------------GUAGGACGUAUCAGUAAUCCAUAAUCUAUA--UUUCAGCCGUGCCUGCUCUAACCGCCACGGUCUGAUCCGCAAGUACGGCCUGAACAUC ------------------...(((...........)))..........--.(((((((((((.(((((..((((...))))..))...))).)))))).))))).... ( -24.40, z-score = -2.34, R) >droWil1.scaffold_181089 6295508 83 + 12369635 -----------------------CUAAAACUAAAUAUUUCACUUUAUU--UUCUAGCCGCGCCUGCUCCAACCGUCACGGUUUGAUCCGUAAAUAUGGUCUGAACAUU -----------------------.........................--(((..(((((....))((.(((((...))))).))...........)))..))).... ( -10.70, z-score = -0.15, R) >droVir3.scaffold_13047 3200337 99 - 19223366 -------UUCACCAGUGCAAAGAAUUAAUGUGCAAUCUAUUCUUAAAA--CUUUAGCCGUGCCUGCUCAAACCGUCAUGGUUUGAUCCGCAAAUAUGGACUGAACAUU -------......(((...((((((..((.....))..))))))...)--))(((((((((..((((((((((.....)))))))...)))..))))).))))..... ( -23.00, z-score = -2.10, R) >droMoj3.scaffold_6540 14594876 97 - 34148556 -------GUAGCCAAAACAAACAGUUAAUGUGUGCUUUAUU--UAAUA--UUUUAGCCGUGCCUGCUCAAACCGACAUGGUCUGAUCCGCAAAUACGGCCUGAACAUU -------..(((((.(((.....)))....)).))).....--.....--.((((((((((..((((((.(((.....))).)))...)))..))))).))))).... ( -19.50, z-score = -0.52, R) >droGri2.scaffold_14830 3983407 91 + 6267026 -------------CGUGCUAUUAAU-AAUAAGCAGCGU-UUGUUAAAA--UUUCAGCCGUGCCUGCUCAAACCGACAUGGUCUGAUUCGCAAAUACGGACUGAACAUC -------------...........(-(((((((...))-))))))...--.((((((((((..((((((.(((.....))).)))...)))..))))).))))).... ( -22.90, z-score = -1.77, R) >anoGam1.chr2R 56165199 96 - 62725911 ------------AUGUAGCAGGAUUGUAACCAAAAUUCUGUUUCCGUUCGUUUCAGCCGUGCCUGCUCCAACAACCACGGAAUGAUCCGAAAGUACGGUCUGAACAUU ------------(((.(((((((((........)))))))))..)))..(((.(((((((((.((......))....((((....))))...)))))).))))))... ( -25.70, z-score = -1.90, R) >apiMel3.GroupUn 366775155 107 - 399230636 AUGAUAGUAAGAUAAUAACCUCAAUGUUACAGUUAAUCAUAUUGUUAUU-UUAUAGUCGUGCUUGCGCAAAUAGACAUGGAUUAAUUCGCAAAUAUGGUUUAAAUAUU .((((.((((.((..........)).)))).))))((((((((((((.(-((((.(((.(((....)))....)))))))).)))).....))))))))......... ( -16.10, z-score = 0.48, R) >triCas2.ChLG7 15009212 78 + 17478683 -----------------------------UUAGCAACAAUUUAAUUUAU-UUCCAGCCGAGCUUGCUCAAACAGGCACGGAUUGAUCCGUAAAUAUGGGCUCAAUAUC -----------------------------....................-......(((.(((((......))))).)))((((((((((....))))).)))))... ( -17.70, z-score = -1.82, R) >consensus ______________AUGCAAACAAUCUAACUAUGAUUUAUUAUCUAAU__UUUCAGCCGUGCCUGCUCUAACCGCCACGGUCUGAUCCGCAAGUACGGCCUGAACAUC .......................................................((((((..(((.(.....)....((......)))))..))))))......... ( -7.49 = -7.27 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:01:24 2011