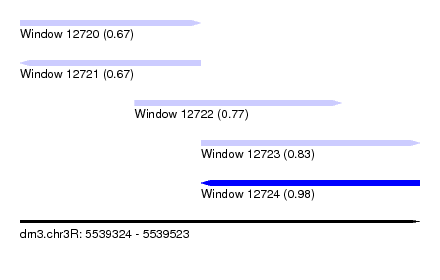
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 5,539,324 – 5,539,523 |
| Length | 199 |
| Max. P | 0.977456 |

| Location | 5,539,324 – 5,539,414 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 52.88 |
| Shannon entropy | 0.95118 |
| G+C content | 0.42372 |
| Mean single sequence MFE | -22.60 |
| Consensus MFE | -6.84 |
| Energy contribution | -5.22 |
| Covariance contribution | -1.62 |
| Combinations/Pair | 2.00 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.670298 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
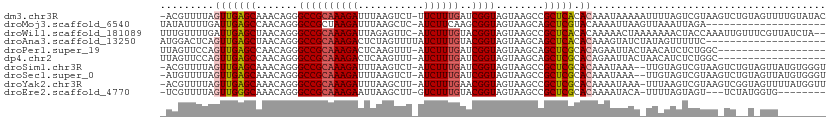
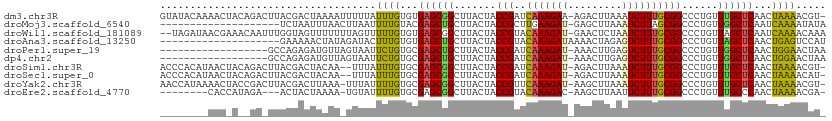
>dm3.chr3R 5539324 90 + 27905053 --------CGUCCAAGCGUUG-GGAAAGCGUCGACACUGUAA-----ACAGUGAGUGUAGACACUGACCU-CAUAACGCUAGAUUAACUUAAAACUGAGAC--GUUA-- --------(((((((((((((-....((.(((..(((((...-----.)))))((((....)))))))))-..)))))))((.....))......)).)))--)...-- ( -24.60, z-score = -1.67, R) >droGri2.scaffold_15074 5202943 95 - 7742996 --------CCAUUUUGCGCUGGACACACA-AUGACGUUCUUUUACCCAGUUGACACGUCAUAGUUUAAC--CAUUGCGCCAGAUUAAGUGUAAAUCGAAAAA-GUUA-- --------..((((.(((((((....((.-(((((((((............)).))))))).))....)--))..)))).))))..................-....-- ( -18.40, z-score = -0.74, R) >droMoj3.scaffold_6540 33950339 94 + 34148556 --------CUAUUCUGCGCUUGACAAAUACGCGACGCUUUUUAGUCUGAAGCAUUUGUGUUUGCUGA----UGCUGCGCUAAAUUAAAUCAAAAUCAAAAAG-GUUA-- --------.......((((((..((((((((.((.(((((.......))))).))))))))))..))----....)))).............((((.....)-))).-- ( -16.90, z-score = 0.80, R) >droVir3.scaffold_12822 1335258 98 + 4096053 ------CUUAAUGUCGCGCUCGACAAGACGACGACAUUGUCCAAA--AACGUAAAUGUCCUCGUUUGUUGUCACGGCGCUAAAUUAAUUCCAAAUCGAGAAA-GUUA-- ------.(((((...(((((.((((((((((.((((((.......--......)))))).)))))..)))))..)))))...)))))...............-....-- ( -25.42, z-score = -2.13, R) >droWil1.scaffold_181089 9377356 96 + 12369635 --------AAUAUUUGCAUCU-GUUUAUCGUCGACAUUGCCCACA--AACAUCAAUGUCGAGGUUACAUC-UUUGGUGCUAGAUUAAUUUUAAAUCGGAAAC-AUGUUA --------.......((((((-((....(.(((((((((......--.....))))))))).)..)))..-...)))))..((((.......))))(....)-...... ( -20.30, z-score = -1.46, R) >droAna3.scaffold_13250 68485 95 + 3535662 ------AUUUUAUUGGCGCCG-AAAAAGUGAAGGCAUUGUAAU----GUAAUAUAUGUCGACAUCAGCCA-AUUGGCGCCAAAUUAAUCUUGAACUGAGGCAAGUUA-- ------......(((((((((-(....(((..(((((.(((..----....))))))))..)))......-.))))))))))..........((((......)))).-- ( -24.10, z-score = -1.46, R) >droPer1.super_19 624857 100 - 1869541 AGCAUAGAAAGAUAAGCGCCC-AGAC-UCGUCGACGUUGCGCACA--GUAUACAACGUCGUGACCA-AUG-GUUGGCGCUAGAUAAAACCAAACU--GAAAC-GCUUUA .............(((((..(-((..-(((.((((((((......--.....)))))))))))...-.((-(((..(....)....)))))..))--)...)-)))).. ( -23.40, z-score = -0.29, R) >dp4.chr2 25266233 100 - 30794189 AGCAUAGAAAGAUAAGCGCCC-AGAC-UCGUCGACGUUGCGCCCA--GUAUACAACGUCGUGACCA-AUG-GUUGGCGCUAGAUAAAACCAAACU--GAAAC-GCUUUA .............(((((..(-((..-(((.((((((((......--.....)))))))))))...-.((-(((..(....)....)))))..))--)...)-)))).. ( -23.40, z-score = -0.49, R) >droSim1.chr3R 11714455 90 - 27517382 --------CGGCUAAGCGUUG-GGAAAGCGUCGACACUGUAA-----ACAGUGAGUGUAGACACUGACCU-CAUAACGCUAGAUUAACUUAAAACUGAGAC--GUUA-- --------((.(((.((((((-....((.(((..(((((...-----.)))))((((....)))))))))-..))))))))).....((((....)))).)--)...-- ( -23.70, z-score = -1.38, R) >droSec1.super_0 4755136 88 - 21120651 --------CGGCUAAGCGUUG-GGAAAGCGUCGACAC--UAA-----ACAGUGAGUGUAGACACUGACCU-CGUAACGCAAGAUUAACUUAAAACUGAGAC--GUUA-- --------.(((...((((((-.((.((.(((.((((--(..-----......))))).))).))....)-).))))))........((((....))))..--))).-- ( -22.50, z-score = -1.04, R) >droYak2.chr3R 9585985 90 + 28832112 --------CGGCUAAGCGUUG-GGUAAGCGUCGACACUGUAA-----ACAGUGAGUGUAGACACUGACCU-UUCAGCGCUAGAUUAAUUUAAAACUGACAC--GUUA-- --------...(((.((((((-(..(((.(((..(((((...-----.)))))((((....)))))))))-)))))))))))...................--....-- ( -25.60, z-score = -1.39, R) >droEre2.scaffold_4770 1680687 90 - 17746568 --------UGGCUAAGCGUUG-AGGAAGCGUCGACACUGUAA-----GUAGUGCGUGUAGACACUGACCU-AAUAACGCUAGAUUAACUUAAAACUGAGAC--GUUA-- --------...(((.((((((-(((.((.(((.((((.(((.-----....))))))).))).))..)))-..))))))))).....((((....))))..--....-- ( -22.90, z-score = -0.84, R) >consensus ________CGGCUAAGCGCUG_AGAAAGCGUCGACACUGUAA_____ACAGUGAAUGUCGACACUGACCU_CAUAGCGCUAGAUUAACUUAAAACUGAGAA__GUUA__ ...............(((((.............((((.................))))................))))).............................. ( -6.84 = -5.22 + -1.62)
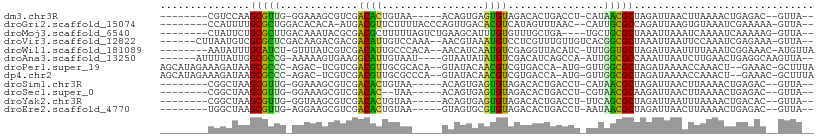
| Location | 5,539,324 – 5,539,414 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 52.88 |
| Shannon entropy | 0.95118 |
| G+C content | 0.42372 |
| Mean single sequence MFE | -22.89 |
| Consensus MFE | -5.22 |
| Energy contribution | -4.68 |
| Covariance contribution | -0.54 |
| Combinations/Pair | 1.64 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.23 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.668332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 5539324 90 - 27905053 --UAAC--GUCUCAGUUUUAAGUUAAUCUAGCGUUAUG-AGGUCAGUGUCUACACUCACUGU-----UUACAGUGUCGACGCUUUCC-CAACGCUUGGACG-------- --...(--(((.((......((.....))(((((..((-.((..((((((......(((((.-----...)))))..))))))..))-)))))))))))))-------- ( -25.20, z-score = -2.24, R) >droGri2.scaffold_15074 5202943 95 + 7742996 --UAAC-UUUUUCGAUUUACACUUAAUCUGGCGCAAUG--GUUAAACUAUGACGUGUCAACUGGGUAAAAGAACGUCAU-UGUGUGUCCAGCGCAAAAUGG-------- --....-......((((.......))))..((((..((--(....((.(((((((.................)))))))-.))....))))))).......-------- ( -20.43, z-score = -0.50, R) >droMoj3.scaffold_6540 33950339 94 - 34148556 --UAAC-CUUUUUGAUUUUGAUUUAAUUUAGCGCAGCA----UCAGCAAACACAAAUGCUUCAGACUAAAAAGCGUCGCGUAUUUGUCAAGCGCAGAAUAG-------- --....-..................((((.((((.((.----...))....((((((((....(((........)))..))))))))...)))).))))..-------- ( -19.50, z-score = -0.83, R) >droVir3.scaffold_12822 1335258 98 - 4096053 --UAAC-UUUCUCGAUUUGGAAUUAAUUUAGCGCCGUGACAACAAACGAGGACAUUUACGUU--UUUGGACAAUGUCGUCGUCUUGUCGAGCGCGACAUUAAG------ --....-.((((......))))(((((...((((..((((((...((((.((((((..((..--..))...)))))).)))).)))))).))))...))))).------ ( -26.90, z-score = -1.79, R) >droWil1.scaffold_181089 9377356 96 - 12369635 UAACAU-GUUUCCGAUUUAAAAUUAAUCUAGCACCAAA-GAUGUAACCUCGACAUUGAUGUU--UGUGGGCAAUGUCGACGAUAAAC-AGAUGCAAAUAUU-------- .....(-((.((.(.((((......((((........)-)))....(.(((((((((.....--......))))))))).).)))))-.)).)))......-------- ( -14.80, z-score = 0.33, R) >droAna3.scaffold_13250 68485 95 - 3535662 --UAACUUGCCUCAGUUCAAGAUUAAUUUGGCGCCAAU-UGGCUGAUGUCGACAUAUAUUAC----AUUACAAUGCCUUCACUUUUU-CGGCGCCAAUAAAAU------ --...((((........))))......((((((((..(-((((....))))).........(----((....)))............-.))))))))......------ ( -20.30, z-score = -1.69, R) >droPer1.super_19 624857 100 + 1869541 UAAAGC-GUUUC--AGUUUGGUUUUAUCUAGCGCCAAC-CAU-UGGUCACGACGUUGUAUAC--UGUGCGCAACGUCGACGA-GUCU-GGGCGCUUAUCUUUCUAUGCU ......-.....--(((.(((........((((((...-.((-(.(((..((((((((....--.....))))))))))).)-))..-.)))))).......))).))) ( -28.76, z-score = -1.16, R) >dp4.chr2 25266233 100 + 30794189 UAAAGC-GUUUC--AGUUUGGUUUUAUCUAGCGCCAAC-CAU-UGGUCACGACGUUGUAUAC--UGGGCGCAACGUCGACGA-GUCU-GGGCGCUUAUCUUUCUAUGCU ......-.....--(((.(((........((((((...-.((-(.(((..((((((((....--.....))))))))))).)-))..-.)))))).......))).))) ( -28.76, z-score = -1.00, R) >droSim1.chr3R 11714455 90 + 27517382 --UAAC--GUCUCAGUUUUAAGUUAAUCUAGCGUUAUG-AGGUCAGUGUCUACACUCACUGU-----UUACAGUGUCGACGCUUUCC-CAACGCUUAGCCG-------- --....--.............((((....(((((..((-.((..((((((......(((((.-----...)))))..))))))..))-)))))))))))..-------- ( -22.70, z-score = -2.14, R) >droSec1.super_0 4755136 88 + 21120651 --UAAC--GUCUCAGUUUUAAGUUAAUCUUGCGUUACG-AGGUCAGUGUCUACACUCACUGU-----UUA--GUGUCGACGCUUUCC-CAACGCUUAGCCG-------- --....--.............(((((....(((((..(-.((..((((((.(((((......-----..)--)))).))))))..))-)))))))))))..-------- ( -22.00, z-score = -1.85, R) >droYak2.chr3R 9585985 90 - 28832112 --UAAC--GUGUCAGUUUUAAAUUAAUCUAGCGCUGAA-AGGUCAGUGUCUACACUCACUGU-----UUACAGUGUCGACGCUUACC-CAACGCUUAGCCG-------- --....--((((((((.(((........))).))))).-.(((.((((((......(((((.-----...)))))..)))))).)))-...))).......-------- ( -23.20, z-score = -1.87, R) >droEre2.scaffold_4770 1680687 90 + 17746568 --UAAC--GUCUCAGUUUUAAGUUAAUCUAGCGUUAUU-AGGUCAGUGUCUACACGCACUAC-----UUACAGUGUCGACGCUUCCU-CAACGCUUAGCCA-------- --....--.............((((....((((((...-(((..((((((.((((.......-----.....)))).)))))).)))-.))))))))))..-------- ( -22.10, z-score = -2.28, R) >consensus __UAAC__GUCUCAAUUUUAAAUUAAUCUAGCGCCAAG_AGGUCAGUGUCGACAUUCACUAU_____UUACAAUGUCGACGCUUUCU_CAACGCUUAGCCG________ ..............................(((((.............((.((((.................)))).))..........)))))............... ( -5.22 = -4.68 + -0.54)
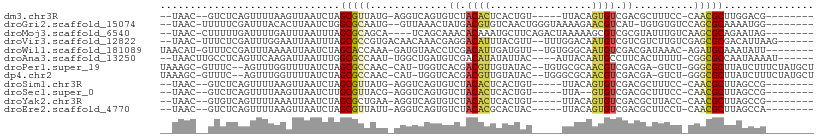

| Location | 5,539,381 – 5,539,484 |
|---|---|
| Length | 103 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 77.62 |
| Shannon entropy | 0.45134 |
| G+C content | 0.41740 |
| Mean single sequence MFE | -25.69 |
| Consensus MFE | -18.74 |
| Energy contribution | -17.56 |
| Covariance contribution | -1.18 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.766503 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 5539381 103 + 27905053 AUAACGCUAGAUUAACUUAAAACUGAGACG---UUAAC-GUUUUAGUUGAGCAAACAGGGCCGCAAAGAUUUAAGUCU-UUCUUUGAUCGGUAGUAAGCCGCUCACAC------- ....................((((((((((---....)-)))))))))((((...(((((....((((((....))))-)))))))...(((.....)))))))....------- ( -26.00, z-score = -1.75, R) >droGri2.scaffold_15074 5203004 105 - 7742996 AUUGCGCCAGAUUAAGUGUAAAUCGAAAAA--GUUAUUGCUUUCGAUUGAGCCAACAGGGCCGUAAAGAUUCAAGCUC-AUCUUUGUACGGUAGUAAGCAGCUCGCAC------- ..(((((........)))))(((((((..(--((....))))))))))((((.......(((((((((((........-)))))..))))))........))))....------- ( -25.36, z-score = -0.71, R) >droMoj3.scaffold_6540 33950399 105 + 34148556 GCUGCGCUAAAUUAAAUCAAAAUCAAAAAG--GUUAUAUAUUUUGAUUGAGCCAACAGGGCCGCUAAGAUUUAAGCUC-AUCUUCAAGCGGUAGUAAGCAGCUCGUAC------- ((((((((.(((((((....((((.....)--)))......))))))).))).......(((((((((((........-)))))..)))))).....)))))......------- ( -27.50, z-score = -2.36, R) >droVir3.scaffold_12822 1335322 105 + 4096053 ACGGCGCUAAAUUAAUUCCAAAUCGAGAAA--GUUAUUUAUUUCGAUUGAGCCAACAGGGCCGCAAGGAUUUAAGUCC-AUCUUUAUGCGGUAGUAAGCAGCUCGCAC------- ...((...............((((((((.(--(....)).))))))))((((.......(((((((((((........-)))))..))))))........))))))..------- ( -25.66, z-score = -1.23, R) >droWil1.scaffold_181089 9377416 114 + 12369635 UUGGUGCUAGAUUAAUUUUAAAUCGGAAACAUGUUAUUUGUUUUGAUUGAGCUAACAGGGCCGCAAAGAUUAGAGUUC-AUCUUUGUACGGUAGUAAGCCGCUCACACAAAAACU ...(((..............(((((.(((((.......))))))))))((((..........((((((((........-))))))))..(((.....))))))).)))....... ( -23.60, z-score = -0.26, R) >droAna3.scaffold_13250 68545 107 + 3535662 UUGGCGCCAAAUUAAUCUUGAACUGAGGCAA-GUUAAUGGACUCAGUUGAGCUAACAGGGCCGCAAAGACUCUAGUUUUAUCUUUGUACGGUAGUAAGCAGCUCACAC------- (((....)))..........(((((((.((.-.....))..)))))))(((((......((((((((((...........))))))..)))).......)))))....------- ( -24.62, z-score = 0.33, R) >droPer1.super_19 624923 105 - 1869541 UUGGCGCUAGAUAAAACC-AAACUGAAACGC-UUUAUUAGUUCCAGUUGAGCCAACAGGGCCGCAAAGACUCAAGUUU-AUCUUUGAUCGGUAGUAAGCAGCUCGCAC------- ...((.............-.(((((.(((..-.......))).)))))((((.......((((((((((.........-.))))))..))))........))))))..------- ( -22.06, z-score = 0.48, R) >dp4.chr2 25266299 105 - 30794189 UUGGCGCUAGAUAAAACC-AAACUGAAACGC-UUUAUUAGUUCCAGUUGAGCCAACAGGGCCGCAAAGACUCAAGUUU-AUCUUUGAUCGGUAGUAAGCAGCUCGCAC------- ...((.............-.(((((.(((..-.......))).)))))((((.......((((((((((.........-.))))))..))))........))))))..------- ( -22.06, z-score = 0.48, R) >droSim1.chr3R 11714512 103 - 27517382 AUAACGCUAGAUUAACUUAAAACUGAGACG---UUAAC-GUUUUAGUUGAGCAAACAGGGCCGCAAAGAUUUAAGUCU-AUCUUUGAUCGGUAGUAAGCCGCUCGCAC------- .....((.............((((((((((---....)-)))))))))((((.......(((((((((((........-)))))))..))))........))))))..------- ( -29.76, z-score = -2.64, R) >droSec1.super_0 4755191 103 - 21120651 GUAACGCAAGAUUAACUUAAAACUGAGACG---UUAAU-GUUUUAGUUGAGCAAACAGGGCCGCAAAGAUUUAAGUCU-AUCUUUGAUCGGUAGUAAGCCGCUCGCAC------- .....((.............((((((((((---....)-)))))))))((((.......(((((((((((........-)))))))..))))........))))))..------- ( -27.66, z-score = -2.00, R) >droYak2.chr3R 9586042 103 + 28832112 UCAGCGCUAGAUUAAUUUAAAACUGACACG---UUAAC-GUUUUAGUUGAGCAAACAGGGCCGCAAAGAUUUAAGCUU-AUCUUUGAACGGUAGUAAGCCGCUCGCAC------- ...((...............((((((.(((---....)-)).))))))((((.......(((((((((((........-)))))))..))))........))))))..------- ( -26.56, z-score = -1.43, R) >droEre2.scaffold_4770 1680744 103 - 17746568 AUAACGCUAGAUUAACUUAAAACUGAGACG---UUAUC-GUUUUAGUUGGGCAAACAGGGCCGCAAAGAAUUAAGCUU-GUCUUUGUACGGUAGUAAGCCGCUCGCAC------- .....((.((..........((((((((((---....)-))))))))).(((..((...((((((((((.........-.))))))..)))).))..))).)).))..------- ( -27.40, z-score = -1.07, R) >consensus AUAGCGCUAGAUUAACUUAAAACUGAGACG___UUAUU_GUUUUAGUUGAGCAAACAGGGCCGCAAAGAUUUAAGUUU_AUCUUUGAACGGUAGUAAGCAGCUCGCAC_______ ....................((((((((............))))))))((((.......((((((((((...........))))))..))))........))))........... (-18.74 = -17.56 + -1.18)
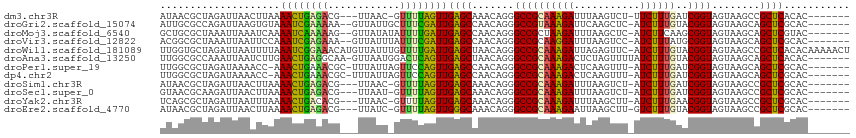
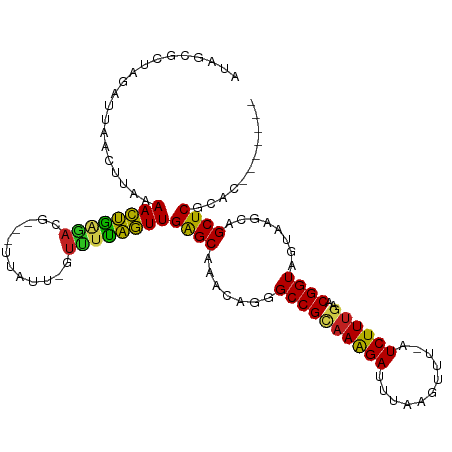
| Location | 5,539,414 – 5,539,523 |
|---|---|
| Length | 109 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 67.84 |
| Shannon entropy | 0.66184 |
| G+C content | 0.40390 |
| Mean single sequence MFE | -21.00 |
| Consensus MFE | -14.01 |
| Energy contribution | -14.11 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.12 |
| Mean z-score | -0.30 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.828173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
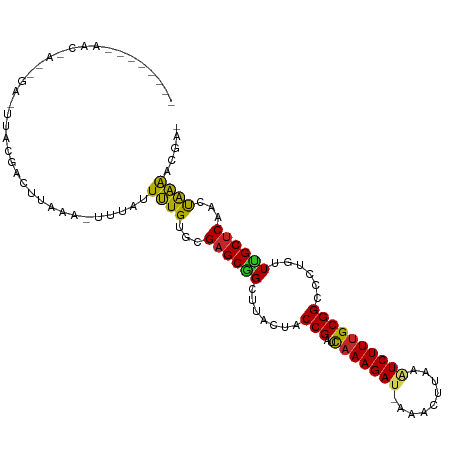
>dm3.chr3R 5539414 109 + 27905053 -ACGUUUUAGUUGAGCAAACAGGGCCGCAAAGAUUUAAGUCU-UUCUUUGAUCGGUAGUAAGCCGCUCACACAAAUAAAAAUUUUAGUCGUAAGUCUGUAGUUUUGUAUAC -........(((((((...(((((....((((((....))))-)))))))...(((.....)))))))).)).....((((((.(((.(....).))).))))))...... ( -22.70, z-score = -1.03, R) >droMoj3.scaffold_6540 33950433 90 + 34148556 UAUAUUUUGAUUGAGCCAACAGGGCCGCUAAGAUUUAAGCUC-AUCUUCAAGCGGUAGUAAGCAGCUCGUACAAAAUUAAGUUAAAUUAGA-------------------- ...((((((..(((((.......(((((((((((........-)))))..))))))........)))))..))))))..............-------------------- ( -20.86, z-score = -1.63, R) >droWil1.scaffold_181089 9377452 108 + 12369635 UUUGUUUUGAUUGAGCUAACAGGGCCGCAAAGAUUAGAGUUC-AUCUUUGUACGGUAGUAAGCCGCUCACACAAAAACUAAAAAAACUACCAAAUUGUUUCGUUAUCUA-- ...(((((...(((((..........((((((((........-))))))))..(((.....))))))))....)))))(((..((((.........))))..)))....-- ( -20.60, z-score = -0.49, R) >droAna3.scaffold_13250 68580 91 + 3535662 AUGGACUCAGUUGAGCUAACAGGGCCGCAAAGACUCUAGUUUUAUCUUUGUACGGUAGUAAGCAGCUCACACAAAGUAUCUAUAGUUUUUC-------------------- (((((((..((((((((......((((((((((...........))))))..)))).......)))))).))..))..)))))........-------------------- ( -19.12, z-score = -0.29, R) >droPer1.super_19 624957 92 - 1869541 UUAGUUCCAGUUGAGCCAACAGGGCCGCAAAGACUCAAGUUU-AUCUUUGAUCGGUAGUAAGCAGCUCGCACAGAAUUACUAACAUCUCUGGC------------------ .((((((..(((((((.......((((((((((.........-.))))))..))))........))))).)).))))))..............------------------ ( -18.06, z-score = 0.52, R) >dp4.chr2 25266333 92 - 30794189 UUAGUUCCAGUUGAGCCAACAGGGCCGCAAAGACUCAAGUUU-AUCUUUGAUCGGUAGUAAGCAGCUCGCACAGAAUUACUAACAUCUCUGGC------------------ .((((((..(((((((.......((((((((((.........-.))))))..))))........))))).)).))))))..............------------------ ( -18.06, z-score = 0.52, R) >droSim1.chr3R 11714545 107 - 27517382 -ACGUUUUAGUUGAGCAAACAGGGCCGCAAAGAUUUAAGUCU-AUCUUUGAUCGGUAGUAAGCCGCUCGCACAAAUAAA--UUGUAGUCGUAAGUCUGUAGUUAUGUGGGU -...........((((.......(((((((((((........-)))))))..))))........)))).((((....((--((((((.(....).)))))))).))))... ( -24.86, z-score = -0.71, R) >droSec1.super_0 4755224 107 - 21120651 -AUGUUUUAGUUGAGCAAACAGGGCCGCAAAGAUUUAAGUCU-AUCUUUGAUCGGUAGUAAGCCGCUCGCACAAAUAAA--UUGUAGUCGUAAGUCUGUAGUUAUGUGGGU -...........((((.......(((((((((((........-)))))))..))))........)))).((((....((--((((((.(....).)))))))).))))... ( -24.86, z-score = -0.93, R) >droYak2.chr3R 9586075 108 + 28832112 -ACGUUUUAGUUGAGCAAACAGGGCCGCAAAGAUUUAAGCUU-AUCUUUGAACGGUAGUAAGCCGCUCGCACAAAAUAAA-UUUAAGUCGUAAGUCGGUAGUUUUAUGGUU -..(((((.(((((((.......(((((((((((........-)))))))..))))........))))).)))))))...-....(..((((((........))))))..) ( -21.96, z-score = 0.13, R) >droEre2.scaffold_4770 1680777 97 - 17746568 -UCGUUUUAGUUGGGCAAACAGGGCCGCAAAGAAUUAAGCUU-GUCUUUGUACGGUAGUAAGCCGCUCGCACAAAAUACA-UUUUAGUAGU---UCUAUGGUG-------- -..(((((.(((((((..........(((((((.........-.)))))))..(((.....)))))))).))))))).((-(..(((....---.)))..)))-------- ( -18.90, z-score = 0.90, R) >consensus _ACGUUUUAGUUGAGCAAACAGGGCCGCAAAGAUUUAAGUUU_AUCUUUGAUCGGUAGUAAGCCGCUCGCACAAAAUAAA_UUAAAGUCGUAA_UC__U_GUU________ .........(((((((.......((((((((((...........))))))..))))........))))).))....................................... (-14.01 = -14.11 + 0.10)
| Location | 5,539,414 – 5,539,523 |
|---|---|
| Length | 109 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 67.84 |
| Shannon entropy | 0.66184 |
| G+C content | 0.40390 |
| Mean single sequence MFE | -19.91 |
| Consensus MFE | -13.05 |
| Energy contribution | -12.10 |
| Covariance contribution | -0.95 |
| Combinations/Pair | 1.40 |
| Mean z-score | -0.95 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.977456 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
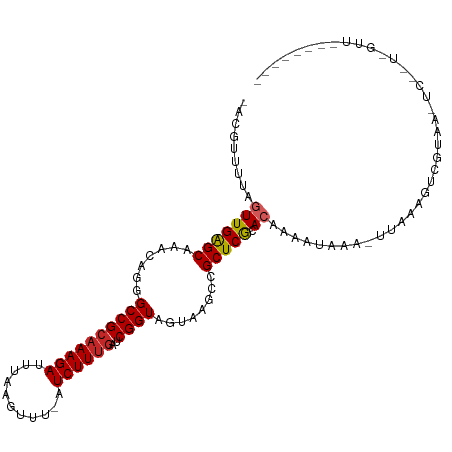
>dm3.chr3R 5539414 109 - 27905053 GUAUACAAAACUACAGACUUACGACUAAAAUUUUUAUUUGUGUGAGCGGCUUACUACCGAUCAAAGAA-AGACUUAAAUCUUUGCGGCCCUGUUUGCUCAACUAAAACGU- ...............................(((((.(((.(..(((((.......(((..((((((.-.........)))))))))..)))))..).))).)))))...- ( -16.50, z-score = 0.07, R) >droMoj3.scaffold_6540 33950433 90 - 34148556 --------------------UCUAAUUUAACUUAAUUUUGUACGAGCUGCUUACUACCGCUUGAAGAU-GAGCUUAAAUCUUAGCGGCCCUGUUGGCUCAAUCAAAAUAUA --------------------..............((((((...(((((....((..(((((..(((((-........))))))))))....)).)))))...))))))... ( -20.70, z-score = -2.25, R) >droWil1.scaffold_181089 9377452 108 - 12369635 --UAGAUAACGAAACAAUUUGGUAGUUUUUUUAGUUUUUGUGUGAGCGGCUUACUACCGUACAAAGAU-GAACUCUAAUCUUUGCGGCCCUGUUAGCUCAAUCAAAACAAA --....(((.(((((.........))))).)))(((((....((((((((......(((..(((((((-........))))))))))....))).)))))...)))))... ( -21.30, z-score = -0.34, R) >droAna3.scaffold_13250 68580 91 - 3535662 --------------------GAAAAACUAUAGAUACUUUGUGUGAGCUGCUUACUACCGUACAAAGAUAAAACUAGAGUCUUUGCGGCCCUGUUAGCUCAACUGAGUCCAU --------------------..............((((.((.(((((((.......(((..(((((((.........))))))))))......))))))))).)))).... ( -19.52, z-score = -1.09, R) >droPer1.super_19 624957 92 + 1869541 ------------------GCCAGAGAUGUUAGUAAUUCUGUGCGAGCUGCUUACUACCGAUCAAAGAU-AAACUUGAGUCUUUGCGGCCCUGUUGGCUCAACUGGAACUAA ------------------.((((........(((......)))(((((....((..(((..(((((((-........))))))))))....)).)))))..))))...... ( -21.60, z-score = -0.25, R) >dp4.chr2 25266333 92 + 30794189 ------------------GCCAGAGAUGUUAGUAAUUCUGUGCGAGCUGCUUACUACCGAUCAAAGAU-AAACUUGAGUCUUUGCGGCCCUGUUGGCUCAACUGGAACUAA ------------------.((((........(((......)))(((((....((..(((..(((((((-........))))))))))....)).)))))..))))...... ( -21.60, z-score = -0.25, R) >droSim1.chr3R 11714545 107 + 27517382 ACCCACAUAACUACAGACUUACGACUACAA--UUUAUUUGUGCGAGCGGCUUACUACCGAUCAAAGAU-AGACUUAAAUCUUUGCGGCCCUGUUUGCUCAACUAAAACGU- ..............................--((((.(((.((((((((.......(((..(((((((-........))))))))))..)))))))).))).))))....- ( -20.40, z-score = -1.80, R) >droSec1.super_0 4755224 107 + 21120651 ACCCACAUAACUACAGACUUACGACUACAA--UUUAUUUGUGCGAGCGGCUUACUACCGAUCAAAGAU-AGACUUAAAUCUUUGCGGCCCUGUUUGCUCAACUAAAACAU- ..............................--((((.(((.((((((((.......(((..(((((((-........))))))))))..)))))))).))).))))....- ( -20.40, z-score = -2.39, R) >droYak2.chr3R 9586075 108 - 28832112 AACCAUAAAACUACCGACUUACGACUUAAA-UUUAUUUUGUGCGAGCGGCUUACUACCGUUCAAAGAU-AAGCUUAAAUCUUUGCGGCCCUGUUUGCUCAACUAAAACGU- ..............................-......(((.((((((((.......(((..(((((((-........))))))))))..)))))))).))).........- ( -18.60, z-score = -0.61, R) >droEre2.scaffold_4770 1680777 97 + 17746568 --------CACCAUAGA---ACUACUAAAA-UGUAUUUUGUGCGAGCGGCUUACUACCGUACAAAGAC-AAGCUUAAUUCUUUGCGGCCCUGUUUGCCCAACUAAAACGA- --------..(((((((---(.(((.....-.)))))))))).).(.(((..((..(((..((((((.-.........)))))))))....))..))))...........- ( -18.50, z-score = -0.58, R) >consensus ________AAC_A__GA_UUACGACUUAAA_UUUAUUUUGUGCGAGCGGCUUACUACCGAUCAAAGAU_AAACUUAAAUCUUUGCGGCCCUGUUUGCUCAACUAAAACGA_ ....................................((((...((((((.......(((..(((((((.........))))))))))......))))))...))))..... (-13.05 = -12.10 + -0.95)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:01:22 2011