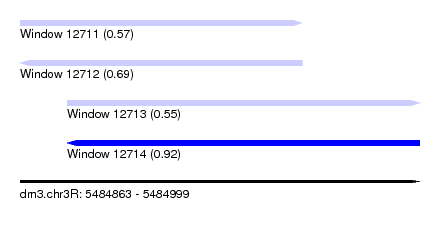
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 5,484,863 – 5,484,999 |
| Length | 136 |
| Max. P | 0.919690 |

| Location | 5,484,863 – 5,484,959 |
|---|---|
| Length | 96 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.73 |
| Shannon entropy | 0.30628 |
| G+C content | 0.52000 |
| Mean single sequence MFE | -29.23 |
| Consensus MFE | -16.88 |
| Energy contribution | -17.20 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.566429 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
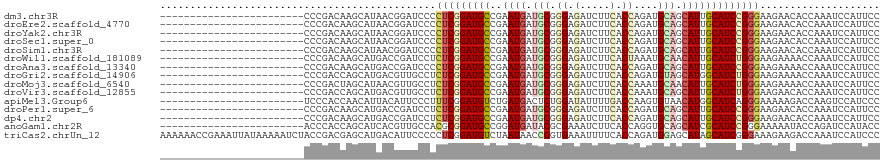
>dm3.chr3R 5484863 96 + 27905053 ------------------------CCCGACAAGCAUAACGGAUCCCCUCGGAUGCCGAAUGAUGCGGGAGAUCUUCACCAGAUGCAGCAUUGCAUCCGGGAAGAACACCAAAUCCAUUCC ------------------------((((.((..(((..(((((((....)))).))).))).)))))).(.(((((.((.(((((......))))).))))))).).............. ( -32.20, z-score = -2.71, R) >droEre2.scaffold_4770 1633388 96 - 17746568 ------------------------CCCGACAAGCAUAACGGAUCCCCUCGGAUGCCGAAUGAUGCGGGAGAUCUUCACCAGAUGCAGCAUUGCAUCCGGGAAGAACACCAAAUCCAUUCC ------------------------((((.((..(((..(((((((....)))).))).))).)))))).(.(((((.((.(((((......))))).))))))).).............. ( -32.20, z-score = -2.71, R) >droYak2.chr3R 9538409 96 + 28832112 ------------------------CCCGACAAGCAUAACGGAUCCCCUCGGAUGCCGAAUGAUGCGGGAGAUCUUCACCAGAUGCAGCAUUGCAUCCGGGAAGAACACCAAAUCCAUUCC ------------------------((((.((..(((..(((((((....)))).))).))).)))))).(.(((((.((.(((((......))))).))))))).).............. ( -32.20, z-score = -2.71, R) >droSec1.super_0 4708327 96 - 21120651 ------------------------CCCGACAAGCAUAACGGAUCCCCUCGGAUGCCGAAUGAUGCGGGAGAUCUUCACCAGAUGCAGCAUUGCAUCCGGGAAGAACACCAAAUCCAUUCC ------------------------((((.((..(((..(((((((....)))).))).))).)))))).(.(((((.((.(((((......))))).))))))).).............. ( -32.20, z-score = -2.71, R) >droSim1.chr3R 11666777 96 - 27517382 ------------------------CCCGACAAGCAUAACGGAUCCCCUCGGAUGCCGAAUGAUGCGGGAGAUCUUCACCAGAUGCAGCAUUGCAUCCGGGAAGAACACCAAAUCCAUUCC ------------------------((((.((..(((..(((((((....)))).))).))).)))))).(.(((((.((.(((((......))))).))))))).).............. ( -32.20, z-score = -2.71, R) >droWil1.scaffold_181089 5684636 96 - 12369635 ------------------------CCCGACAAGCAUGACCGAUCCUCUCGGAUGCCGAAUGAUGCGGGAGAUCUUCACUAAAUGCAACAUUGCAUCUGGGAAGAAAACCAAAUCCAUUCC ------------------------........((((..((((.....)))))))).(((((((...((...(((((.(((.(((((....))))).))))))))...))..)).))))). ( -26.30, z-score = -1.82, R) >droAna3.scaffold_13340 9548841 96 - 23697760 ------------------------CCCGACAAGCAUGACCGAUCCCCUCGGAUGCCGAAUGAUGCGGGAGAUCUUCACCAGAUGCAGCAUUGCAUCCGGGAAGAAAACCAAAUCCAUUCC ------------------------........((((..((((.....)))))))).(((((((...((...(((((.((.(((((......))))).)))))))...))..)).))))). ( -29.80, z-score = -2.00, R) >droGri2.scaffold_14906 5736962 96 - 14172833 ------------------------CCCGACCAGCAUGACGUUGCCUCUCGGAUGCCGAAUGAUGCGGGAGAUCUUCACCAGAUGUAGCAUGGCAUCUGGGAAGAAAACCAAAUCCAUUCC ------------------------.((((...(((......)))...)))).....(((((((...((...(((((.((((((((......)))))))))))))...))..)).))))). ( -33.20, z-score = -2.28, R) >droMoj3.scaffold_6540 5887283 96 + 34148556 ------------------------CCCGACUAGCAUAACGUUGCCUCUCGGAUGCCGAAUGAUGCGGGAGAUCUUCACCAAAUGCAACAUUGCAUCUGGGAAGAAAACCAAAUCCAUUCC ------------------------.((((...(((......)))...)))).....(((((((...((...(((((.(((.(((((....))))).))))))))...))..)).))))). ( -29.50, z-score = -2.85, R) >droVir3.scaffold_12855 5564876 96 + 10161210 ------------------------CCCGACCAGCAUGACGUUGCCUCUCGGAUGCCGAAUGAUGCGGGAGAUCUUCACCAAAUGCAGCAUUGCAUCUGGGAAGAACACCAAAUCCAUUCC ------------------------.((((...(((......)))...)))).....(((((((...((.(.(((((.(((.((((......)))).)))))))).).))..)).))))). ( -30.30, z-score = -1.80, R) >apiMel3.Group6 2769334 96 + 14581788 ------------------------UCCCACCAACAUUACAUUCCCUUUCGGAUGUCUGAUGACUCUGGAUAUUUUGACCAAGUGUAACAUGGCAUCAGGGAAAAAGACCAAGUCCAUCCC ------------------------.............((((((......))))))..(((((((.(((...((((..((..((((......))))..))..))))..)))))).)))).. ( -19.90, z-score = 0.34, R) >droPer1.super_6 1037762 96 + 6141320 ------------------------CCCGACAAGCAUGACCGAUCCUCUCGGAUGCCGAAUGAUGCGGGAGAUCUUCACCAGAUGCAGCAUUGCAUCCGGGAAGAACACCAAAUCCAUUCC ------------------------........((((..((((.....)))))))).(((((((...((.(.(((((.((.(((((......))))).))))))).).))..)).))))). ( -31.00, z-score = -2.11, R) >dp4.chr2 25694906 96 + 30794189 ------------------------CCCGACAAGCAUGACCGAUCCUCUCGGAUGCCGAAUGAUGCGGGAGAUCUUCACCAGAUGCAGCAUUGCAUCCGGGAAGAACACCAAAUCCAUUCC ------------------------........((((..((((.....)))))))).(((((((...((.(.(((((.((.(((((......))))).))))))).).))..)).))))). ( -31.00, z-score = -2.11, R) >anoGam1.chr2R 34966291 96 + 62725911 ------------------------ACCCACCAGCAUCACGUUGCCACGCGGAUGCCGGAUGAUACGCGAAAUCUUCACCAGGUGCAGCAUCGCAUCCGGGAAAAAUACCAGAUCCAUACC ------------------------.(((....(((((.(((......)))))))).(((((....((...((((.....))))...))....)))))))).................... ( -26.80, z-score = -1.39, R) >triCas2.chrUn_12 219541 120 + 931581 AAAAAACCGAAAUUAUAAAAAUCUACCGACGAGCAUGACAUUCCCCCUCGGAUGUCUAAUAACCCGUGAAAUUUUCACCAGAUGGAGCAUAGCAUCGGGAAAGAAGACCAAAUCCAUCCC .....................(((............(((((((......)))))))......(((((((.....))))..((((........)))))))..)))................ ( -19.70, z-score = -0.22, R) >consensus ________________________CCCGACAAGCAUGACGGAUCCCCUCGGAUGCCGAAUGAUGCGGGAGAUCUUCACCAGAUGCAGCAUUGCAUCCGGGAAGAACACCAAAUCCAUUCC ..............................................(((((((((..((((.(((.((.(.....).))....))).))))))))))))).................... (-16.88 = -17.20 + 0.32)
| Location | 5,484,863 – 5,484,959 |
|---|---|
| Length | 96 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.73 |
| Shannon entropy | 0.30628 |
| G+C content | 0.52000 |
| Mean single sequence MFE | -35.52 |
| Consensus MFE | -26.54 |
| Energy contribution | -26.72 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.691895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 5484863 96 - 27905053 GGAAUGGAUUUGGUGUUCUUCCCGGAUGCAAUGCUGCAUCUGGUGAAGAUCUCCCGCAUCAUUCGGCAUCCGAGGGGAUCCGUUAUGCUUGUCGGG------------------------ ................((((((((((((((....))))))))).)))))...((((((.(((.(((.((((....)))))))..)))..)).))))------------------------ ( -37.40, z-score = -1.73, R) >droEre2.scaffold_4770 1633388 96 + 17746568 GGAAUGGAUUUGGUGUUCUUCCCGGAUGCAAUGCUGCAUCUGGUGAAGAUCUCCCGCAUCAUUCGGCAUCCGAGGGGAUCCGUUAUGCUUGUCGGG------------------------ ................((((((((((((((....))))))))).)))))...((((((.(((.(((.((((....)))))))..)))..)).))))------------------------ ( -37.40, z-score = -1.73, R) >droYak2.chr3R 9538409 96 - 28832112 GGAAUGGAUUUGGUGUUCUUCCCGGAUGCAAUGCUGCAUCUGGUGAAGAUCUCCCGCAUCAUUCGGCAUCCGAGGGGAUCCGUUAUGCUUGUCGGG------------------------ ................((((((((((((((....))))))))).)))))...((((((.(((.(((.((((....)))))))..)))..)).))))------------------------ ( -37.40, z-score = -1.73, R) >droSec1.super_0 4708327 96 + 21120651 GGAAUGGAUUUGGUGUUCUUCCCGGAUGCAAUGCUGCAUCUGGUGAAGAUCUCCCGCAUCAUUCGGCAUCCGAGGGGAUCCGUUAUGCUUGUCGGG------------------------ ................((((((((((((((....))))))))).)))))...((((((.(((.(((.((((....)))))))..)))..)).))))------------------------ ( -37.40, z-score = -1.73, R) >droSim1.chr3R 11666777 96 + 27517382 GGAAUGGAUUUGGUGUUCUUCCCGGAUGCAAUGCUGCAUCUGGUGAAGAUCUCCCGCAUCAUUCGGCAUCCGAGGGGAUCCGUUAUGCUUGUCGGG------------------------ ................((((((((((((((....))))))))).)))))...((((((.(((.(((.((((....)))))))..)))..)).))))------------------------ ( -37.40, z-score = -1.73, R) >droWil1.scaffold_181089 5684636 96 + 12369635 GGAAUGGAUUUGGUUUUCUUCCCAGAUGCAAUGUUGCAUUUAGUGAAGAUCUCCCGCAUCAUUCGGCAUCCGAGAGGAUCGGUCAUGCUUGUCGGG------------------------ .((((((....((...((((((.(((((((....))))))).).)))))...))....))))))(((((((((.....))))..))))).......------------------------ ( -29.10, z-score = -0.51, R) >droAna3.scaffold_13340 9548841 96 + 23697760 GGAAUGGAUUUGGUUUUCUUCCCGGAUGCAAUGCUGCAUCUGGUGAAGAUCUCCCGCAUCAUUCGGCAUCCGAGGGGAUCGGUCAUGCUUGUCGGG------------------------ .((((((....((...((((((((((((((....))))))))).)))))...))....))))))(((((((((.....))))..))))).......------------------------ ( -37.70, z-score = -1.73, R) >droGri2.scaffold_14906 5736962 96 + 14172833 GGAAUGGAUUUGGUUUUCUUCCCAGAUGCCAUGCUACAUCUGGUGAAGAUCUCCCGCAUCAUUCGGCAUCCGAGAGGCAACGUCAUGCUGGUCGGG------------------------ .((((((....((...((((((((((((........))))))).)))))...))....)))))).....((((..((((......))))..)))).------------------------ ( -35.60, z-score = -1.85, R) >droMoj3.scaffold_6540 5887283 96 - 34148556 GGAAUGGAUUUGGUUUUCUUCCCAGAUGCAAUGUUGCAUUUGGUGAAGAUCUCCCGCAUCAUUCGGCAUCCGAGAGGCAACGUUAUGCUAGUCGGG------------------------ .((((((....((...((((((((((((((....))))))))).)))))...))....)))))).....((((..((((......))))..)))).------------------------ ( -36.60, z-score = -3.08, R) >droVir3.scaffold_12855 5564876 96 - 10161210 GGAAUGGAUUUGGUGUUCUUCCCAGAUGCAAUGCUGCAUUUGGUGAAGAUCUCCCGCAUCAUUCGGCAUCCGAGAGGCAACGUCAUGCUGGUCGGG------------------------ .((((((....((.(.((((((((((((((....))))))))).))))).).))....)))))).....((((..((((......))))..)))).------------------------ ( -38.50, z-score = -2.37, R) >apiMel3.Group6 2769334 96 - 14581788 GGGAUGGACUUGGUCUUUUUCCCUGAUGCCAUGUUACACUUGGUCAAAAUAUCCAGAGUCAUCAGACAUCCGAAAGGGAAUGUAAUGUUGGUGGGA------------------------ ((((.((((...))))...))))....(((((((((((.((((.........)))).(((....))).(((....)))..))))))).))))....------------------------ ( -26.20, z-score = 0.14, R) >droPer1.super_6 1037762 96 - 6141320 GGAAUGGAUUUGGUGUUCUUCCCGGAUGCAAUGCUGCAUCUGGUGAAGAUCUCCCGCAUCAUUCGGCAUCCGAGAGGAUCGGUCAUGCUUGUCGGG------------------------ .((((((....((.(.((((((((((((((....))))))))).))))).).))....))))))(((((((((.....))))..))))).......------------------------ ( -38.30, z-score = -1.91, R) >dp4.chr2 25694906 96 - 30794189 GGAAUGGAUUUGGUGUUCUUCCCGGAUGCAAUGCUGCAUCUGGUGAAGAUCUCCCGCAUCAUUCGGCAUCCGAGAGGAUCGGUCAUGCUUGUCGGG------------------------ .((((((....((.(.((((((((((((((....))))))))).))))).).))....))))))(((((((((.....))))..))))).......------------------------ ( -38.30, z-score = -1.91, R) >anoGam1.chr2R 34966291 96 - 62725911 GGUAUGGAUCUGGUAUUUUUCCCGGAUGCGAUGCUGCACCUGGUGAAGAUUUCGCGUAUCAUCCGGCAUCCGCGUGGCAACGUGAUGCUGGUGGGU------------------------ ((((((((......(((((..((((.((((....)))).))))..))))).)).))))))..((((((((.((((....)))))))))))).....------------------------ ( -35.20, z-score = -0.85, R) >triCas2.chrUn_12 219541 120 - 931581 GGGAUGGAUUUGGUCUUCUUUCCCGAUGCUAUGCUCCAUCUGGUGAAAAUUUCACGGGUUAUUAGACAUCCGAGGGGGAAUGUCAUGCUCGUCGGUAGAUUUUUAUAAUUUCGGUUUUUU .(((((((..((((..((......)).))))...))))))).(((((((((((((((((.....(((((((.....)).)))))..)))))).))..))))))))).............. ( -30.30, z-score = 0.17, R) >consensus GGAAUGGAUUUGGUGUUCUUCCCGGAUGCAAUGCUGCAUCUGGUGAAGAUCUCCCGCAUCAUUCGGCAUCCGAGAGGAUCCGUCAUGCUUGUCGGG________________________ .((((((....((...((((((((((((........))))))).)))))...))....)))))).....((((.(((..........))).))))......................... (-26.54 = -26.72 + 0.18)
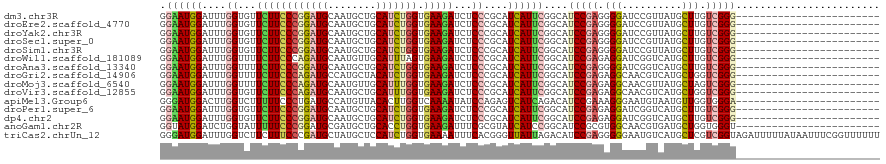
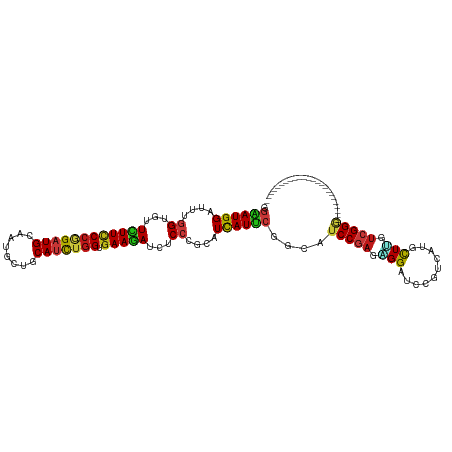
| Location | 5,484,879 – 5,484,999 |
|---|---|
| Length | 120 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.19 |
| Shannon entropy | 0.32117 |
| G+C content | 0.50222 |
| Mean single sequence MFE | -38.30 |
| Consensus MFE | -19.05 |
| Energy contribution | -19.36 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.46 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.552906 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
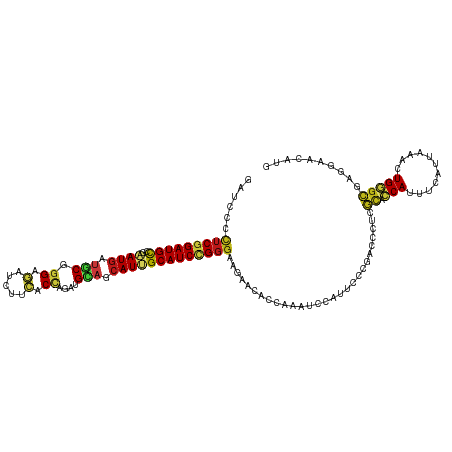
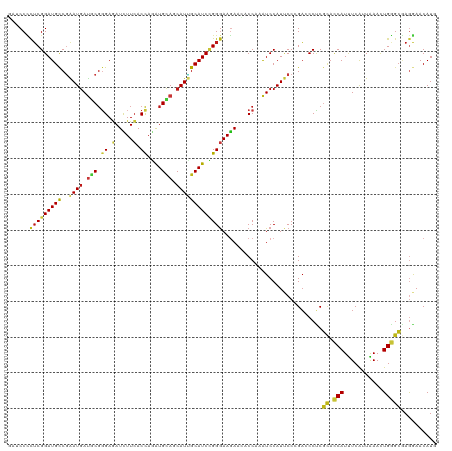
>dm3.chr3R 5484879 120 + 27905053 GAUCCCCUCGGAUGCCGAAUGAUGCGGGAGAUCUUCACCAGAUGCAGCAUUGCAUCCGGGAAGAACACCAAAUCCAUUCCCGACCCUCGCACCAUUUCAUUAAACUGGGCGAGGAACAUG .......((((.....(((((((...((.(.(((((.((.(((((......))))).))))))).).))..)).))))))))).((((((.(((.((.....)).)))))))))...... ( -40.70, z-score = -2.72, R) >droEre2.scaffold_4770 1633404 120 - 17746568 GAUCCCCUCGGAUGCCGAAUGAUGCGGGAGAUCUUCACCAGAUGCAGCAUUGCAUCCGGGAAGAACACCAAAUCCAUUCCCGACCCUCGCACCAUUUCAUUAAACUGGGCGAGGAACAUG .......((((.....(((((((...((.(.(((((.((.(((((......))))).))))))).).))..)).))))))))).((((((.(((.((.....)).)))))))))...... ( -40.70, z-score = -2.72, R) >droYak2.chr3R 9538425 120 + 28832112 GAUCCCCUCGGAUGCCGAAUGAUGCGGGAGAUCUUCACCAGAUGCAGCAUUGCAUCCGGGAAGAACACCAAAUCCAUUCCCGACCCUCGCACCAUUUCAUUAAACUGGGCGAGGAACAUG .......((((.....(((((((...((.(.(((((.((.(((((......))))).))))))).).))..)).))))))))).((((((.(((.((.....)).)))))))))...... ( -40.70, z-score = -2.72, R) >droSec1.super_0 4708343 120 - 21120651 GAUCCCCUCGGAUGCCGAAUGAUGCGGGAGAUCUUCACCAGAUGCAGCAUUGCAUCCGGGAAGAACACCAAAUCCAUUCCCGACCCUCGCACCAUUUCAUUAAACUGGGCGAGGAACAUG .......((((.....(((((((...((.(.(((((.((.(((((......))))).))))))).).))..)).))))))))).((((((.(((.((.....)).)))))))))...... ( -40.70, z-score = -2.72, R) >droSim1.chr3R 11666793 120 - 27517382 GAUCCCCUCGGAUGCCGAAUGAUGCGGGAGAUCUUCACCAGAUGCAGCAUUGCAUCCGGGAAGAACACCAAAUCCAUUCCCGACCCUCGCACCAUUUCAUUAAACUGGGCGAGGAACAUG .......((((.....(((((((...((.(.(((((.((.(((((......))))).))))))).).))..)).))))))))).((((((.(((.((.....)).)))))))))...... ( -40.70, z-score = -2.72, R) >droWil1.scaffold_181089 5684652 120 - 12369635 GAUCCUCUCGGAUGCCGAAUGAUGCGGGAGAUCUUCACUAAAUGCAACAUUGCAUCUGGGAAGAAAACCAAAUCCAUUCCAGAGCCCCUCACCAUUUCAUUAAAUUGGGCAAGGAACAUG ..((.((((((((((..((((.((((..((.......))...)))).)))))))))))))).))............((((...((((...................))))..)))).... ( -31.01, z-score = -0.81, R) >droAna3.scaffold_13340 9548857 120 - 23697760 GAUCCCCUCGGAUGCCGAAUGAUGCGGGAGAUCUUCACCAGAUGCAGCAUUGCAUCCGGGAAGAAAACCAAAUCCAUUCCCGACCCUCGCACCAUCUCGUUAAACUGGGCGAGGAACAUG .......((((.....(((((((...((...(((((.((.(((((......))))).)))))))...))..)).))))))))).((((((.(((...........)))))))))...... ( -39.40, z-score = -1.98, R) >droGri2.scaffold_14906 5736978 120 - 14172833 UUGCCUCUCGGAUGCCGAAUGAUGCGGGAGAUCUUCACCAGAUGUAGCAUGGCAUCUGGGAAGAAAACCAAAUCCAUUCCGGAUCCUCGCACCAUCUCAUUGAAUUGGGCGAGGAACAUA ....((((((.((........)).)))))).(((((.((((((((......)))))))))))))........(((.....)))(((((((.(((..((...))..))))))))))..... ( -46.50, z-score = -3.18, R) >droMoj3.scaffold_6540 5887299 120 + 34148556 UUGCCUCUCGGAUGCCGAAUGAUGCGGGAGAUCUUCACCAAAUGCAACAUUGCAUCUGGGAAGAAAACCAAAUCCAUUCCGGAGCCUCGCACCAUCUCAUUGAAUUGGGCGAGAAACAUA ..((((((((.((........)).)))))).(((((.(((.(((((....))))).))))))))........(((.....)))))(((((.(((..((...))..))))))))....... ( -38.30, z-score = -2.09, R) >droVir3.scaffold_12855 5564892 120 + 10161210 UUGCCUCUCGGAUGCCGAAUGAUGCGGGAGAUCUUCACCAAAUGCAGCAUUGCAUCUGGGAAGAACACCAAAUCCAUUCCGGAGCCUCGCACCAUCUCAUUGAAUUGGGCGAGAAACAUA ....(((.(((((((........)))((.(.(((((.(((.((((......)))).)))))))).).))........))))))).(((((.(((..((...))..))))))))....... ( -38.40, z-score = -1.59, R) >apiMel3.Group6 2769350 120 + 14581788 UUCCCUUUCGGAUGUCUGAUGACUCUGGAUAUUUUGACCAAGUGUAACAUGGCAUCAGGGAAAAAGACCAAGUCCAUCCCAGAUCCUCUUUGCAUUUCGUUGAAUUGUGAAAGAAACAUU ....((((((((.(((....))))))..(((.((..((.((((((((...((.(((.((((....(((...)))..)))).)))))...)))))))).))..)).))))))))....... ( -31.70, z-score = -1.20, R) >droPer1.super_6 1037778 120 + 6141320 GAUCCUCUCGGAUGCCGAAUGAUGCGGGAGAUCUUCACCAGAUGCAGCAUUGCAUCCGGGAAGAACACCAAAUCCAUUCCAGAUCCUCGCACCAUUUCAUUGAACUGGGCGAGGAACAUA .((((....))))...(((((((...((.(.(((((.((.(((((......))))).))))))).).))..)).)))))..(.(((((((.(((.(((...))).)))))))))).)... ( -44.70, z-score = -3.38, R) >dp4.chr2 25694922 120 + 30794189 GAUCCUCUCGGAUGCCGAAUGAUGCGGGAGAUCUUCACCAGAUGCAGCAUUGCAUCCGGGAAGAACACCAAAUCCAUUCCAGAUCCUCGCACCAUUUCAUUGAACUGGGCGAGGAACAUA .((((....))))...(((((((...((.(.(((((.((.(((((......))))).))))))).).))..)).)))))..(.(((((((.(((.(((...))).)))))))))).)... ( -44.70, z-score = -3.38, R) >anoGam1.chr2R 34966307 120 + 62725911 UUGCCACGCGGAUGCCGGAUGAUACGCGAAAUCUUCACCAGGUGCAGCAUCGCAUCCGGGAAAAAUACCAGAUCCAUACCGGUACCGCGAAUCAUUUCGUUAAACUGUACCAAAAACAUC ......(((((.(((((((((....((((...((.(((...))).))..))))(((.((........)).))).))).)))))))))))............................... ( -32.50, z-score = -1.49, R) >triCas2.chrUn_12 219581 120 + 931581 UUCCCCCUCGGAUGUCUAAUAACCCGUGAAAUUUUCACCAGAUGGAGCAUAGCAUCGGGAAAGAAGACCAAAUCCAUCCCUGAACCCCGCACCAUUUCGUUGAAUUGUGCCAAAAACAUG .........((((((((.....(((((((.....))))..((((........))))))).....))))...)))).............((((.((((....)))).)))).......... ( -23.80, z-score = -0.56, R) >consensus GAUCCCCUCGGAUGCCGAAUGAUGCGGGAGAUCUUCACCAGAUGCAGCAUUGCAUCCGGGAAGAACACCAAAUCCAUUCCCGACCCUCGCACCAUUUCAUUAAACUGGGCGAGGAACAUG ......(((((((((..((((.(((.((.(.....).))....))).)))))))))))))............................((.(((...........))))).......... (-19.05 = -19.36 + 0.31)
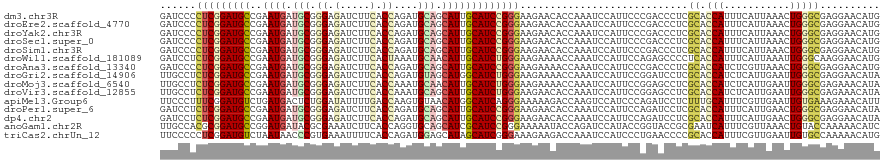
| Location | 5,484,879 – 5,484,999 |
|---|---|
| Length | 120 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.19 |
| Shannon entropy | 0.32117 |
| G+C content | 0.50222 |
| Mean single sequence MFE | -44.59 |
| Consensus MFE | -31.90 |
| Energy contribution | -31.09 |
| Covariance contribution | -0.81 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.919690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
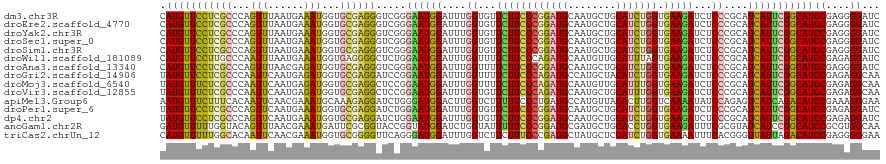
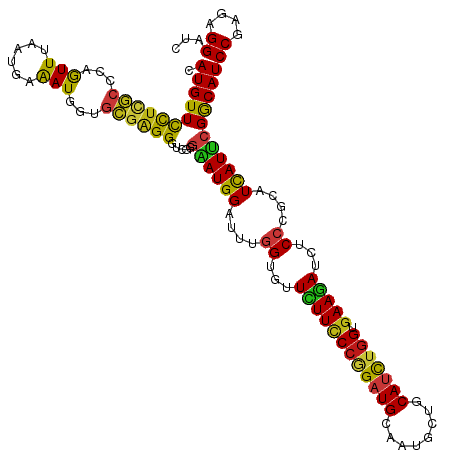
>dm3.chr3R 5484879 120 - 27905053 CAUGUUCCUCGCCCAGUUUAAUGAAAUGGUGCGAGGGUCGGGAAUGGAUUUGGUGUUCUUCCCGGAUGCAAUGCUGCAUCUGGUGAAGAUCUCCCGCAUCAUUCGGCAUCCGAGGGGAUC ......(((((((((...........))).))))))((((((....(((.(((.(.((((((((((((((....))))))))).)))))...)))).))).))))))((((....)))). ( -48.40, z-score = -2.21, R) >droEre2.scaffold_4770 1633404 120 + 17746568 CAUGUUCCUCGCCCAGUUUAAUGAAAUGGUGCGAGGGUCGGGAAUGGAUUUGGUGUUCUUCCCGGAUGCAAUGCUGCAUCUGGUGAAGAUCUCCCGCAUCAUUCGGCAUCCGAGGGGAUC ......(((((((((...........))).))))))((((((....(((.(((.(.((((((((((((((....))))))))).)))))...)))).))).))))))((((....)))). ( -48.40, z-score = -2.21, R) >droYak2.chr3R 9538425 120 - 28832112 CAUGUUCCUCGCCCAGUUUAAUGAAAUGGUGCGAGGGUCGGGAAUGGAUUUGGUGUUCUUCCCGGAUGCAAUGCUGCAUCUGGUGAAGAUCUCCCGCAUCAUUCGGCAUCCGAGGGGAUC ......(((((((((...........))).))))))((((((....(((.(((.(.((((((((((((((....))))))))).)))))...)))).))).))))))((((....)))). ( -48.40, z-score = -2.21, R) >droSec1.super_0 4708343 120 + 21120651 CAUGUUCCUCGCCCAGUUUAAUGAAAUGGUGCGAGGGUCGGGAAUGGAUUUGGUGUUCUUCCCGGAUGCAAUGCUGCAUCUGGUGAAGAUCUCCCGCAUCAUUCGGCAUCCGAGGGGAUC ......(((((((((...........))).))))))((((((....(((.(((.(.((((((((((((((....))))))))).)))))...)))).))).))))))((((....)))). ( -48.40, z-score = -2.21, R) >droSim1.chr3R 11666793 120 + 27517382 CAUGUUCCUCGCCCAGUUUAAUGAAAUGGUGCGAGGGUCGGGAAUGGAUUUGGUGUUCUUCCCGGAUGCAAUGCUGCAUCUGGUGAAGAUCUCCCGCAUCAUUCGGCAUCCGAGGGGAUC ......(((((((((...........))).))))))((((((....(((.(((.(.((((((((((((((....))))))))).)))))...)))).))).))))))((((....)))). ( -48.40, z-score = -2.21, R) >droWil1.scaffold_181089 5684652 120 + 12369635 CAUGUUCCUUGCCCAAUUUAAUGAAAUGGUGAGGGGCUCUGGAAUGGAUUUGGUUUUCUUCCCAGAUGCAAUGUUGCAUUUAGUGAAGAUCUCCCGCAUCAUUCGGCAUCCGAGAGGAUC ...((((((..((..((((....))))))..))))))....((((((....((...((((((.(((((((....))))))).).)))))...))....))))))...((((....)))). ( -38.40, z-score = -1.24, R) >droAna3.scaffold_13340 9548857 120 + 23697760 CAUGUUCCUCGCCCAGUUUAACGAGAUGGUGCGAGGGUCGGGAAUGGAUUUGGUUUUCUUCCCGGAUGCAAUGCUGCAUCUGGUGAAGAUCUCCCGCAUCAUUCGGCAUCCGAGGGGAUC ......(((((((((...........))).))))))((((((....(((.(((...((((((((((((((....))))))))).)))))....))).))).))))))((((....)))). ( -48.20, z-score = -1.99, R) >droGri2.scaffold_14906 5736978 120 + 14172833 UAUGUUCCUCGCCCAAUUCAAUGAGAUGGUGCGAGGAUCCGGAAUGGAUUUGGUUUUCUUCCCAGAUGCCAUGCUACAUCUGGUGAAGAUCUCCCGCAUCAUUCGGCAUCCGAGAGGCAA ..(((..((((((((.(((...))).))).))..(((((((((...(((.(((...((((((((((((........))))))).)))))....))).))).))))).)))))))..))). ( -47.20, z-score = -3.03, R) >droMoj3.scaffold_6540 5887299 120 - 34148556 UAUGUUUCUCGCCCAAUUCAAUGAGAUGGUGCGAGGCUCCGGAAUGGAUUUGGUUUUCUUCCCAGAUGCAAUGUUGCAUUUGGUGAAGAUCUCCCGCAUCAUUCGGCAUCCGAGAGGCAA ..(((((((((((((.(((...))).))).))..((..(((((...(((.(((...((((((((((((((....))))))))).)))))....))).))).)))))...)))))))))). ( -47.40, z-score = -3.24, R) >droVir3.scaffold_12855 5564892 120 - 10161210 UAUGUUUCUCGCCCAAUUCAAUGAGAUGGUGCGAGGCUCCGGAAUGGAUUUGGUGUUCUUCCCAGAUGCAAUGCUGCAUUUGGUGAAGAUCUCCCGCAUCAUUCGGCAUCCGAGAGGCAA ..(((((((((((((.(((...))).))).))..((..(((((...(((.(((.(.((((((((((((((....))))))))).)))))...)))).))).)))))...)))))))))). ( -47.80, z-score = -2.91, R) >apiMel3.Group6 2769350 120 - 14581788 AAUGUUUCUUUCACAAUUCAACGAAAUGCAAAGAGGAUCUGGGAUGGACUUGGUCUUUUUCCCUGAUGCCAUGUUACACUUGGUCAAAAUAUCCAGAGUCAUCAGACAUCCGAAAGGGAA ....(..(((((......................(((((.((((.((((...))))...)))).)).((((.(.....).)))).......)))...(((....)))....)))))..). ( -26.10, z-score = 0.97, R) >droPer1.super_6 1037778 120 - 6141320 UAUGUUCCUCGCCCAGUUCAAUGAAAUGGUGCGAGGAUCUGGAAUGGAUUUGGUGUUCUUCCCGGAUGCAAUGCUGCAUCUGGUGAAGAUCUCCCGCAUCAUUCGGCAUCCGAGAGGAUC .....((((((((((.(((...))).))).)))))))....((((((....((.(.((((((((((((((....))))))))).))))).).))....))))))...((((....)))). ( -51.80, z-score = -3.86, R) >dp4.chr2 25694922 120 - 30794189 UAUGUUCCUCGCCCAGUUCAAUGAAAUGGUGCGAGGAUCUGGAAUGGAUUUGGUGUUCUUCCCGGAUGCAAUGCUGCAUCUGGUGAAGAUCUCCCGCAUCAUUCGGCAUCCGAGAGGAUC .....((((((((((.(((...))).))).)))))))....((((((....((.(.((((((((((((((....))))))))).))))).).))....))))))...((((....)))). ( -51.80, z-score = -3.86, R) >anoGam1.chr2R 34966307 120 - 62725911 GAUGUUUUUGGUACAGUUUAACGAAAUGAUUCGCGGUACCGGUAUGGAUCUGGUAUUUUUCCCGGAUGCGAUGCUGCACCUGGUGAAGAUUUCGCGUAUCAUCCGGCAUCCGCGUGGCAA .......(((.(((........(((....)))((((..((((.((((((.(((.(((((..((((.((((....)))).))))..))))).))).)).))))))))...))))))).))) ( -36.80, z-score = -0.29, R) >triCas2.chrUn_12 219581 120 - 931581 CAUGUUUUUGGCACAAUUCAACGAAAUGGUGCGGGGUUCAGGGAUGGAUUUGGUCUUCUUUCCCGAUGCUAUGCUCCAUCUGGUGAAAAUUUCACGGGUUAUUAGACAUCCGAGGGGGAA ................((((.(.(.((((.(((.(((...((((.(((........))).))))...))).))).)))).).)))))..(..(.(((((........))))).)..)... ( -31.40, z-score = 0.57, R) >consensus CAUGUUCCUCGCCCAGUUUAAUGAAAUGGUGCGAGGGUCGGGAAUGGAUUUGGUGUUCUUCCCGGAUGCAAUGCUGCAUCUGGUGAAGAUCUCCCGCAUCAUUCGGCAUCCGAGAGGAUC .(((((((((((...(((......)))...)))))).....((((((....((...((((((((((((........))))))).)))))...))....)))))))))))((....))... (-31.90 = -31.09 + -0.81)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:01:14 2011