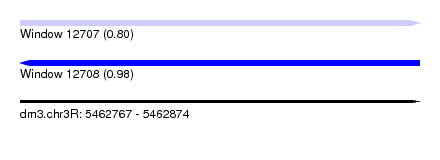
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 5,462,767 – 5,462,874 |
| Length | 107 |
| Max. P | 0.983872 |

| Location | 5,462,767 – 5,462,874 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 60.90 |
| Shannon entropy | 0.73530 |
| G+C content | 0.60116 |
| Mean single sequence MFE | -38.16 |
| Consensus MFE | -14.60 |
| Energy contribution | -17.72 |
| Covariance contribution | 3.12 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.797462 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 5462767 107 + 27905053 GUGGAUGUGGAGGAGGCGGAUGCGGAUGCGGAGGAGGUGGCAUUCGUACUGGCCCCUGGCGCAGCAGAAUUGCUCCCGCUACUGGUACUGCUAUCACGUCCGCCCGA ((((((((((((.((.(((.(((((((((.(......).))))))))))))(((..(((((.(((......)))..)))))..))).)).)).)))))))))).... ( -45.60, z-score = -0.80, R) >droSec1.super_0 4685928 101 - 21120651 GUGGAUGUGGAGGAGGCGGAUGCGGA------GGAGGUGGCAUUCGUACUGGCUCCUGCCGCAGCAGAAUUGCUCCCGCUGCUGUUGCUGCUAUCACGUCCGCCUGA (((((((((((((((.(((.((((((------.(......).))))))))).)))))((.(((((((....((....))..))))))).))..)))))))))).... ( -48.00, z-score = -2.29, R) >droYak2.chr3R 9515806 95 + 28832112 GUGGAUGUGGAGGAGGCGGA------------GGUGGUGGCAUUCGUACUGGCUCCUGCCGCAGCAGAAUUGCUCCCGCUGCUGGUGCUGCUAUCACGUCCGCCGGA ..............((((((------------.(((((((((...((((((((....)))(((((.((.....))..))))).)))))))))))))).))))))... ( -46.30, z-score = -2.08, R) >droEre2.scaffold_4770 1611209 95 - 17746568 GUGGAUGUGGAGGAGGCGGA------------GGAUGUGGCAUUCGUACUGGCUCCUGCCGCAGCAGAAUUGCUCCCGCUGCUGUUGCUGCUAUCACGUCCGCCCGA (((((((((((((((.(((.------------(((((...)))))...))).)))))((.(((((((....((....))..))))))).))..)))))))))).... ( -44.80, z-score = -2.51, R) >droAna3.scaffold_13340 9521547 89 - 23697760 GUGGAUGAGGAGUUAGCGUUUGGGUU------GGAGGCGUUGUUUCCAUUG-CCACUGGUGUUG--GAAUUGGUGCCGUUGCUGUCGCGACCCCCUGA--------- ((((....((((.((((((((.....------..)))))))).))))....-)))).((..((.--.....))..))(((((....))))).......--------- ( -26.60, z-score = 0.17, R) >anoGam1.chr2R 34899704 88 + 62725911 --------ACUGCUGACGGACGA---------AUUGUUGCCGCCGGUACCA--UCCGAUAGCCGCUUGAUAGAUUUCGCUACUACUGUAUGCGUGACACCCUUUGUG --------...((((.((((...---------..((.((((...)))).))--)))).))))(((...........((((((....))).)))...........))) ( -17.65, z-score = 0.29, R) >consensus GUGGAUGUGGAGGAGGCGGAUG__________GGAGGUGGCAUUCGUACUGGCUCCUGCCGCAGCAGAAUUGCUCCCGCUGCUGUUGCUGCUAUCACGUCCGCCCGA ((((((((((.((.((((................(((.(.((.......)).).)))...(((((.(........).)))))...)))).)).)))))))))).... (-14.60 = -17.72 + 3.12)


| Location | 5,462,767 – 5,462,874 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 60.90 |
| Shannon entropy | 0.73530 |
| G+C content | 0.60116 |
| Mean single sequence MFE | -32.85 |
| Consensus MFE | -13.74 |
| Energy contribution | -17.13 |
| Covariance contribution | 3.39 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.983872 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
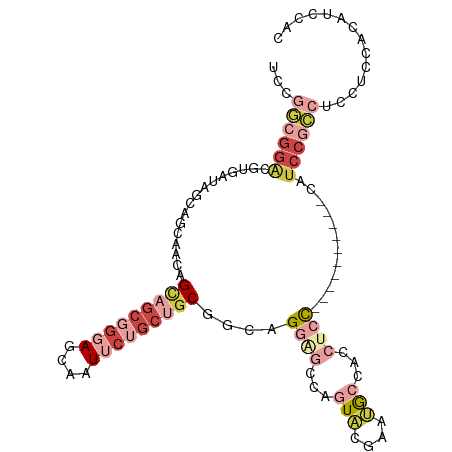
>dm3.chr3R 5462767 107 - 27905053 UCGGGCGGACGUGAUAGCAGUACCAGUAGCGGGAGCAAUUCUGCUGCGCCAGGGGCCAGUACGAAUGCCACCUCCUCCGCAUCCGCAUCCGCCUCCUCCACAUCCAC ..(((((((.(((((.((.......(((((((((....)))))))))...((((((..........))).))).....)))).))).)))))))............. ( -38.40, z-score = -1.10, R) >droSec1.super_0 4685928 101 + 21120651 UCAGGCGGACGUGAUAGCAGCAACAGCAGCGGGAGCAAUUCUGCUGCGGCAGGAGCCAGUACGAAUGCCACCUCC------UCCGCAUCCGCCUCCUCCACAUCCAC ..(((((((.((....)).((....(((((((((....)))))))))((.(((((...(((....)))...))))------))))).)))))))............. ( -40.90, z-score = -2.96, R) >droYak2.chr3R 9515806 95 - 28832112 UCCGGCGGACGUGAUAGCAGCACCAGCAGCGGGAGCAAUUCUGCUGCGGCAGGAGCCAGUACGAAUGCCACCACC------------UCCGCCUCCUCCACAUCCAC ...((((((.(((...(((......(((((((((....)))))))))(((....)))........)))...))).------------)))))).............. ( -39.80, z-score = -3.15, R) >droEre2.scaffold_4770 1611209 95 + 17746568 UCGGGCGGACGUGAUAGCAGCAACAGCAGCGGGAGCAAUUCUGCUGCGGCAGGAGCCAGUACGAAUGCCACAUCC------------UCCGCCUCCUCCACAUCCAC ..(((((((.(.(((.(..(((...(((((((((....)))))))))(((....)))........)))..)))))------------)))))))............. ( -37.70, z-score = -2.33, R) >droAna3.scaffold_13340 9521547 89 + 23697760 ---------UCAGGGGGUCGCGACAGCAACGGCACCAAUUC--CAACACCAGUGG-CAAUGGAAACAACGCCUCC------AACCCAAACGCUAACUCCUCAUCCAC ---------..(((((...(((...((....))........--........(.((-(..((....))..))).).------........)))...)))))....... ( -19.40, z-score = -0.36, R) >anoGam1.chr2R 34899704 88 - 62725911 CACAAAGGGUGUCACGCAUACAGUAGUAGCGAAAUCUAUCAAGCGGCUAUCGGA--UGGUACCGGCGGCAACAAU---------UCGUCCGUCAGCAGU-------- ......((((....(((.(((....))))))..)))).....((.(((..((((--((((....))(....)...---------.))))))..))).))-------- ( -20.90, z-score = -0.25, R) >consensus UCCGGCGGACGUGAUAGCAGCAACAGCAGCGGGAGCAAUUCUGCUGCGGCAGGAGCCAGUACGAAUGCCACCUCC__________CAUCCGCCUCCUCCACAUCCAC ...((((((................(((((((((....)))))))))....((((...(((....)))...))))............)))))).............. (-13.74 = -17.13 + 3.39)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:01:08 2011