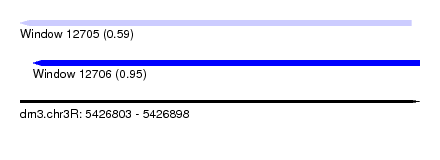
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 5,426,803 – 5,426,898 |
| Length | 95 |
| Max. P | 0.954656 |

| Location | 5,426,803 – 5,426,896 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 68.85 |
| Shannon entropy | 0.56961 |
| G+C content | 0.42151 |
| Mean single sequence MFE | -23.05 |
| Consensus MFE | -9.74 |
| Energy contribution | -9.42 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.589740 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
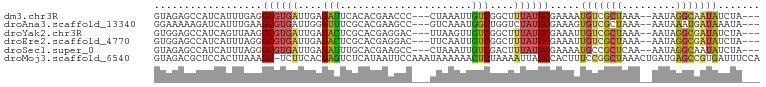
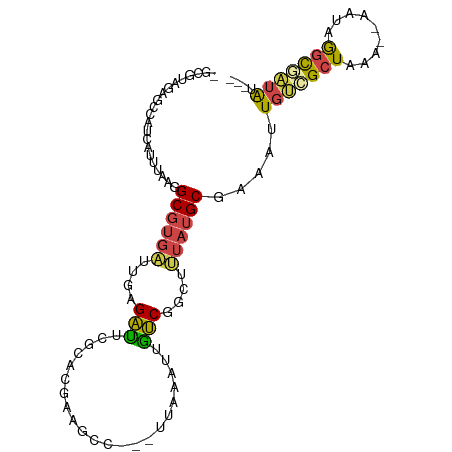
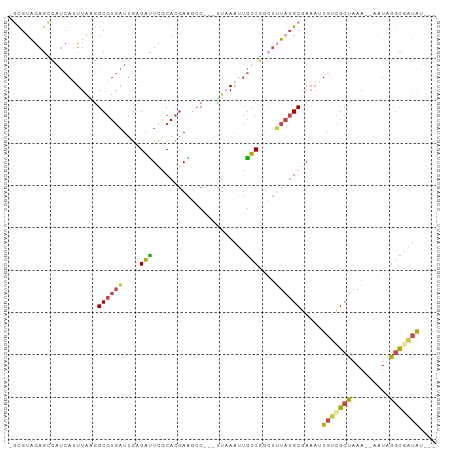
>dm3.chr3R 5426803 93 - 27905053 GUAGAGCCAUCAUUUGAGGCGUGAUUGAGAUUCACACGAACCC---CUAAAUUGUCGGCUUUAUGCGAAAAUGUCGCUAAA--AAUAGGCAAUAUCUA--- ((((((((..((....(((((((..((.....))))))....)---))....))..))))))))((((.....))))....--...............--- ( -21.70, z-score = -1.20, R) >droAna3.scaffold_13340 17544177 93 + 23697760 GGAAAAAGAUCAUUUGAAGCGUGAUUGGGUUUCGCACGAAGCC---GUCAAAUGGCUGGUCUAUGCGAAAGUGUCGCUAAA--AAUAAAUGAUAAAUA--- ........((((((((....(((((..(.(((((((...((((---((...))))))......))))))).))))))....--..)))))))).....--- ( -23.50, z-score = -1.87, R) >droYak2.chr3R 9479167 93 - 28832112 GUGGAGCCAUCAGUUAAGGCGUGAUUGAGACUCGCACGAGGAC---UUAAGUUGUCGGCUUUAUGCGAAAUUGUCGCUAAA--AAUAGGCGAUAUCUA--- ((((((((.(((((((.....)))))))..(((....)))(((---.......))))))))))).......(((((((...--....)))))))....--- ( -27.00, z-score = -1.34, R) >droEre2.scaffold_4770 1575044 93 + 17746568 GUGGAGCCAUCAUUUAGGGCGUGAUUGAGACUCGCACGAGGAC---UUCAAUUGUCGGCUUUAUGCGAAAUUGUCGCUAAA--AAUAGGCGAUAUCUA--- ((((((((..(.....)(((..(((((((.(((....)))...---)))))))))))))))))).......(((((((...--....)))))))....--- ( -24.90, z-score = -0.73, R) >droSec1.super_0 4650210 93 + 21120651 GUAGAGCCAUCAUUUAGGGCGUGAUUGAGAUUUGCACGAAGCC---CUAAAUUGUCGACUUUAUGCGAAAAUGCCGCUCAA--AAUAGGCAAUAUCUA--- (((((((.((.((((((((((((...........)))...)))---)))))).)).).)))))).......((((......--....)))).......--- ( -22.30, z-score = -0.84, R) >droMoj3.scaffold_6540 13195225 100 - 34148556 GUAGACGCUCCACUUAAAGC-UCUUCACGAGUCUCAUAAUUCCAAAUAAAAAACUCUAAAAUUAGCCACUUUCCGGCUAAACUGAUGAGCCGUGAUUUCCA ..(((.(((........)))-)))(((((.(.(((((.........((........))...((((((.......))))))....)))))))))))...... ( -18.90, z-score = -2.42, R) >consensus GUAGAGCCAUCAUUUAAGGCGUGAUUGAGAUUCGCACGAAGCC___UUAAAUUGUCGGCUUUAUGCGAAAUUGUCGCUAAA__AAUAGGCGAUAUCUA___ ..................((((((....(((......................)))....)))))).....(((((((.........)))))))....... ( -9.74 = -9.42 + -0.33)
| Location | 5,426,806 – 5,426,898 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 68.28 |
| Shannon entropy | 0.57605 |
| G+C content | 0.43446 |
| Mean single sequence MFE | -26.97 |
| Consensus MFE | -9.74 |
| Energy contribution | -9.42 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.56 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.954656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
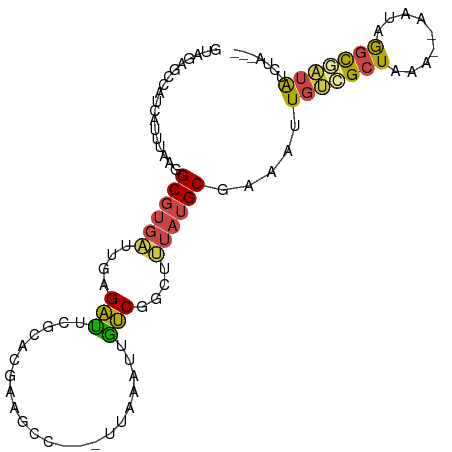
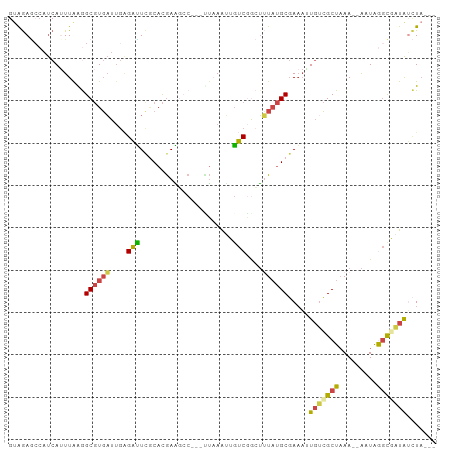
>dm3.chr3R 5426806 92 - 27905053 -GCGUAGAGCCAUCAUUUGAGGCGUGAUUGAGAUUCACACGAACCC---CUAAAUUGUCGGCUUUAUGCGAAAAUGUCGCUAAA--AAUAGGCAAUAU--- -((((((((((..((....(((((((..((.....))))))....)---))....))..)))))))))).....((((......--....))))....--- ( -26.30, z-score = -2.47, R) >droAna3.scaffold_13340 17544180 93 + 23697760 GCUGGAAAAAGAUCAUUUGAAGCGUGAUUGGGUUUCGCACGAAGCC---GUCAAAUGGCUGGUCUAUGCGAAAGUGUCGCUAAA--AAUAAAUGAUAA--- ...........((((((((....(((((..(.(((((((...((((---((...))))))......))))))).))))))....--..))))))))..--- ( -23.50, z-score = -1.26, R) >droYak2.chr3R 9479170 92 - 28832112 -GCGUGGAGCCAUCAGUUAAGGCGUGAUUGAGACUCGCACGAGGAC---UUAAGUUGUCGGCUUUAUGCGAAAUUGUCGCUAAA--AAUAGGCGAUAU--- -((((((((((.(((((((.....)))))))..(((....)))(((---.......))))))))))))).....(((((((...--....))))))).--- ( -33.30, z-score = -3.28, R) >droEre2.scaffold_4770 1575047 92 + 17746568 -GCGUGGAGCCAUCAUUUAGGGCGUGAUUGAGACUCGCACGAGGAC---UUCAAUUGUCGGCUUUAUGCGAAAUUGUCGCUAAA--AAUAGGCGAUAU--- -((((((((((..(.....)(((..(((((((.(((....)))...---)))))))))))))))))))).....(((((((...--....))))))).--- ( -31.20, z-score = -2.57, R) >droSec1.super_0 4650213 92 + 21120651 -GCGUAGAGCCAUCAUUUAGGGCGUGAUUGAGAUUUGCACGAAGCC---CUAAAUUGUCGACUUUAUGCGAAAAUGCCGCUCAA--AAUAGGCAAUAU--- -(((((((((.((.((((((((((((...........)))...)))---)))))).)).).)))))))).....((((......--....))))....--- ( -28.60, z-score = -2.68, R) >droMoj3.scaffold_6540 13195228 99 - 34148556 -UCGUAGACGCUCCACUUAAAGC-UCUUCACGAGUCUCAUAAUUCCAAAUAAAAAACUCUAAAAUUAGCCACUUUCCGGCUAAACUGAUGAGCCGUGAUUU -....(((.(((........)))-)))(((((.(.(((((.........((........))...((((((.......))))))....)))))))))))... ( -18.90, z-score = -2.19, R) >consensus _GCGUAGAGCCAUCAUUUAAGGCGUGAUUGAGAUUCGCACGAAGCC___UUAAAUUGUCGGCUUUAUGCGAAAUUGUCGCUAAA__AAUAGGCGAUAU___ .....................((((((....(((......................)))....)))))).....(((((((.........))))))).... ( -9.74 = -9.42 + -0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:01:07 2011