
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 5,299,550 – 5,299,657 |
| Length | 107 |
| Max. P | 0.971020 |

| Location | 5,299,550 – 5,299,657 |
|---|---|
| Length | 107 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 69.84 |
| Shannon entropy | 0.58130 |
| G+C content | 0.46657 |
| Mean single sequence MFE | -26.76 |
| Consensus MFE | -18.07 |
| Energy contribution | -18.24 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.971020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
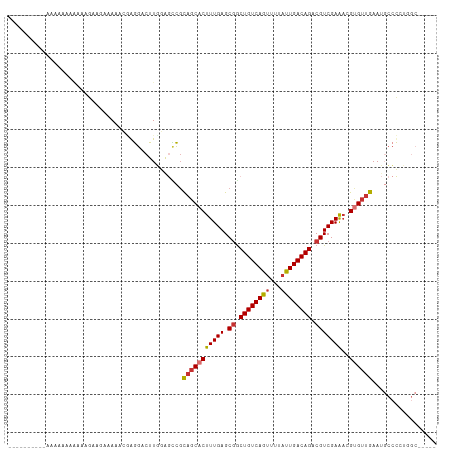
>dm3.chr3R 5299550 107 + 27905053 -AAAAAAAAAAAAACCAAAAUGAAGAAAAACGAGGACUUGGAGCCGCAGCACUUUGAGCGGCUGUCAGUUUUAUUGACAGACGUCGAAACGUGUUGAAUGCCCCUAGC----- -...................................((.((.((..(((((((((((.((.((((((((...)))))))).)))))))..))))))...)).)).)).----- ( -26.40, z-score = -1.78, R) >droSim1.chr3R 11482763 97 - 27517382 -----------UGCAAAAAAAGAUGGAAAACCGGGACUUGGAGCCGCAGCACUUUGAGCGGCUGUCAGUUUUAUUGACAGACGUCGAGACGUGUUGAAUGCCCUUGGC----- -----------.((.........(((....)))...(..((.((..(((((((((..(((.((((((((...)))))))).)))..))).))))))...))))..)))----- ( -27.70, z-score = -0.60, R) >droSec1.super_0 4523742 108 - 21120651 UGCACAAAAAAAAAGCAAAAAGAUGGAAAACCGGGACUUGGAGACGCAGCACUUUGAGCGGCUGUCAGUUUUAUUGACAGACGUCGAGACGUGUUGAAUGCCCCUGGC----- (((...........))).............(((((....(....).(((((((((..(((.((((((((...)))))))).)))..))).))))))......))))).----- ( -30.50, z-score = -1.36, R) >droYak2.chr3R 9348339 101 + 28832112 -------CGAAAAAACAGAAGGAAAGAAAUCGAGGACUUGGAGCCGCAGCACUUUGAGCGGCUGUCAGUUUUAUUGACAGACGUCGAGACGCGUUGAAUGCCCCUGGC----- -------........(((..((.....(((((((((((.(.((((((..........)))))).).)))))).(((((....)))))..)).))).....)).)))..----- ( -24.80, z-score = 0.45, R) >droEre2.scaffold_4770 1442143 100 - 17746568 -------UGCAAAAAAAGAAGGAGC-AAAUGGAGGACUUGGAGCCGCAGCACUUUGAGCGGCUGUCAGUUUUAUUGACAGACGUCGAGACGCGUUGAAUGCCCCUGGC----- -------............(((.((-(((((.((((((.(.((((((..........)))))).).)))))).(((((....)))))....))))...))).)))...----- ( -26.90, z-score = 0.22, R) >droAna3.scaffold_13340 18949605 104 - 23697760 -------CGCAAAAAAAAAAAAGUACAAAAUGGGGGCUUCAGGUCCU--CACUUUGAGCAACUGUCAGUUUUUCUGACAGACGUCGAAUCGUGUUAAAUGCCCAGCGCGAGAG -------(((............((.((((.(((((((.....)))))--)).)))).))..(((((((.....)))))))..).))..(((((((........)))))))... ( -32.80, z-score = -2.94, R) >dp4.chr2 28982276 82 - 30794189 --------------------CGCAGAAAGG-AAAGACUUUAAAUUGCAGCACUUUGAGCGGCUGUCAGUUUUAUUGACAGACGUCGAAACGUGUUGAAAUAGU---------- --------------------.((((((((.-.....))))...))))((((((((((.((.((((((((...)))))))).)))))))..)))))........---------- ( -22.50, z-score = -1.77, R) >droPer1.super_7 2479364 82 - 4445127 --------------------CGCAGAAAGG-AAAGACUUUAAAUUGCAGCACUUUGAGCGGCUGUCAGUUUUAUUGACAGACGUCGAAACGUGUUGAAAUAGU---------- --------------------.((((((((.-.....))))...))))((((((((((.((.((((((((...)))))))).)))))))..)))))........---------- ( -22.50, z-score = -1.77, R) >consensus __________AAAAAAAAAAAGAAGAAAAACGAGGACUUGGAGCCGCAGCACUUUGAGCGGCUGUCAGUUUUAUUGACAGACGUCGAAACGUGUUGAAUGCCCCUGGC_____ ..............................................(((((((((((.((.((((((((...)))))))).)))))))..))))))................. (-18.07 = -18.24 + 0.17)

| Location | 5,299,550 – 5,299,657 |
|---|---|
| Length | 107 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 69.84 |
| Shannon entropy | 0.58130 |
| G+C content | 0.46657 |
| Mean single sequence MFE | -21.57 |
| Consensus MFE | -13.39 |
| Energy contribution | -13.25 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.37 |
| Mean z-score | -0.74 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.791441 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 5299550 107 - 27905053 -----GCUAGGGGCAUUCAACACGUUUCGACGUCUGUCAAUAAAACUGACAGCCGCUCAAAGUGCUGCGGCUCCAAGUCCUCGUUUUUCUUCAUUUUGGUUUUUUUUUUUUU- -----(((.(((((...((.(((.(((.((((.((((((.......)))))).)).)))))))).))..))))).)))..................................- ( -24.20, z-score = -1.13, R) >droSim1.chr3R 11482763 97 + 27517382 -----GCCAAGGGCAUUCAACACGUCUCGACGUCUGUCAAUAAAACUGACAGCCGCUCAAAGUGCUGCGGCUCCAAGUCCCGGUUUUCCAUCUUUUUUUGCA----------- -----((...((((.......((((....)))).(((((.......)))))(((((..........))))).....)))).((....))..........)).----------- ( -23.20, z-score = -0.65, R) >droSec1.super_0 4523742 108 + 21120651 -----GCCAGGGGCAUUCAACACGUCUCGACGUCUGUCAAUAAAACUGACAGCCGCUCAAAGUGCUGCGUCUCCAAGUCCCGGUUUUCCAUCUUUUUGCUUUUUUUUUGUGCA -----(((((((((.......((((....)))).(((((.......)))))((.((.......)).))........)))))((....))..................)).)). ( -23.70, z-score = -0.19, R) >droYak2.chr3R 9348339 101 - 28832112 -----GCCAGGGGCAUUCAACGCGUCUCGACGUCUGUCAAUAAAACUGACAGCCGCUCAAAGUGCUGCGGCUCCAAGUCCUCGAUUUCUUUCCUUCUGUUUUUUCG------- -----..(..((((.......((((....)))).(((((.......)))))(((((..........))))).....))))..).......................------- ( -22.50, z-score = -0.11, R) >droEre2.scaffold_4770 1442143 100 + 17746568 -----GCCAGGGGCAUUCAACGCGUCUCGACGUCUGUCAAUAAAACUGACAGCCGCUCAAAGUGCUGCGGCUCCAAGUCCUCCAUUU-GCUCCUUCUUUUUUUGCA------- -----...(((((((......((((....)))).(((((.......)))))(((((..........)))))...............)-))))))............------- ( -25.50, z-score = -0.75, R) >droAna3.scaffold_13340 18949605 104 + 23697760 CUCUCGCGCUGGGCAUUUAACACGAUUCGACGUCUGUCAGAAAAACUGACAGUUGCUCAAAGUG--AGGACCUGAAGCCCCCAUUUUGUACUUUUUUUUUUUUGCG------- .((((((..((((((......(((......)))(((((((.....))))))).))))))..)))--))).....................................------- ( -25.70, z-score = -1.08, R) >dp4.chr2 28982276 82 + 30794189 ----------ACUAUUUCAACACGUUUCGACGUCUGUCAAUAAAACUGACAGCCGCUCAAAGUGCUGCAAUUUAAAGUCUUU-CCUUUCUGCG-------------------- ----------..........(((.(((.((((.((((((.......)))))).)).))))))))..(((....((((.....-.)))).))).-------------------- ( -13.90, z-score = -0.99, R) >droPer1.super_7 2479364 82 + 4445127 ----------ACUAUUUCAACACGUUUCGACGUCUGUCAAUAAAACUGACAGCCGCUCAAAGUGCUGCAAUUUAAAGUCUUU-CCUUUCUGCG-------------------- ----------..........(((.(((.((((.((((((.......)))))).)).))))))))..(((....((((.....-.)))).))).-------------------- ( -13.90, z-score = -0.99, R) >consensus _____GCCAGGGGCAUUCAACACGUCUCGACGUCUGUCAAUAAAACUGACAGCCGCUCAAAGUGCUGCGGCUCCAAGUCCUCGUUUUCCUUCUUUUUGUUUUU__________ .........((((....((.(((.....((((.((((((.......)))))).)).))...))).))...))))....................................... (-13.39 = -13.25 + -0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:00:57 2011