| Sequence ID | dm3.chr3R |
|---|---|
| Location | 5,284,442 – 5,284,496 |
| Length | 54 |
| Max. P | 0.963345 |

| Location | 5,284,442 – 5,284,496 |
|---|---|
| Length | 54 |
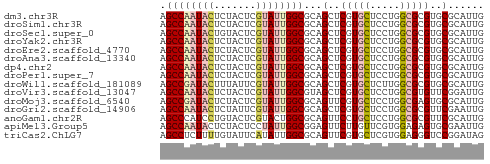
| Sequences | 15 |
| Columns | 54 |
| Reading direction | reverse |
| Mean pairwise identity | 90.09 |
| Shannon entropy | 0.24591 |
| G+C content | 0.57037 |
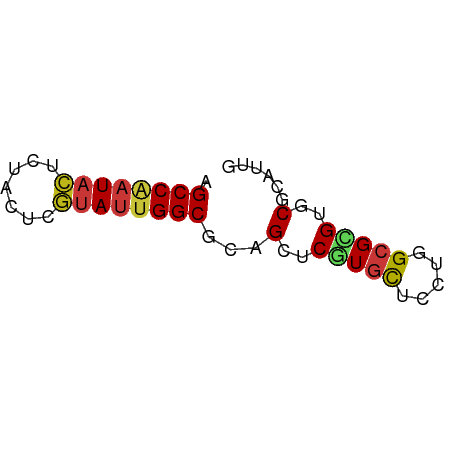
| Mean single sequence MFE | -21.50 |
| Consensus MFE | -14.58 |
| Energy contribution | -14.96 |
| Covariance contribution | 0.38 |
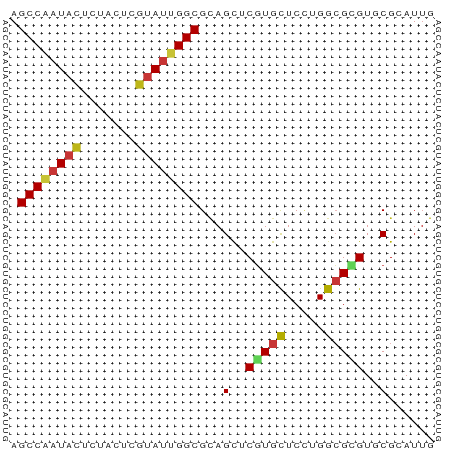
| Combinations/Pair | 1.36 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.963345 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 5284442 54 - 27905053 AGCCAAUACUCUACUCGUAUUGGCGCAGCUCGUGCUCCUGGCGCGUGCGCAUUG .((((((((.......))))))))((.((.(((((.....))))).)))).... ( -25.30, z-score = -3.89, R) >droSim1.chr3R 11467945 54 + 27517382 AGCCAAUACUCUACUCGUAUUGGCGCAGCUCGUGCUCCUGGCGCGUGCGCAUUG .((((((((.......))))))))((.((.(((((.....))))).)))).... ( -25.30, z-score = -3.89, R) >droSec1.super_0 4508791 54 + 21120651 AGCCAAUACUGUACUCGUAUUGGCGCAGCUCGUGCUCCUGGCGCGUGCGCAUUG .((((((((.......))))))))((.((.(((((.....))))).)))).... ( -25.30, z-score = -3.25, R) >droYak2.chr3R 9332865 54 - 28832112 AGCCAAUACUCUACUCGUAUUGGCGCAGCUCGUGCUCCUGGCGCGUGCGCAUUG .((((((((.......))))))))((.((.(((((.....))))).)))).... ( -25.30, z-score = -3.89, R) >droEre2.scaffold_4770 1426806 54 + 17746568 AGCCAAUACUCUACUCGUAUUGGCGCAGCUCGUGCUCCUGGCGCGUGCGCAUUG .((((((((.......))))))))((.((.(((((.....))))).)))).... ( -25.30, z-score = -3.89, R) >droAna3.scaffold_13340 18933195 54 + 23697760 AGCCAAUACUCUACUCGUAUUGGCGCAGCUCGUGCUCCUGGCGCGUGCGCAUUG .((((((((.......))))))))((.((.(((((.....))))).)))).... ( -25.30, z-score = -3.89, R) >dp4.chr2 27661085 54 + 30794189 AGCCAAUACUCUACUCGUAUUGGCGCAGCUCGUGCUCCUGGCGCGUGCGCAUUG .((((((((.......))))))))((.((.(((((.....))))).)))).... ( -25.30, z-score = -3.89, R) >droPer1.super_7 1209342 54 - 4445127 AGCCAAUACUCUACUCGUAUUGGCGCAGCUCGUGCUCCUGGCGCGUGCGCAUUG .((((((((.......))))))))((.((.(((((.....))))).)))).... ( -25.30, z-score = -3.89, R) >droWil1.scaffold_181089 2671006 54 + 12369635 AGCCGAUACUUUAUUCGUAUUGGCGCAGCUCGUGCUCUUGGCGCGUGCGCAUUG .((((((((.......))))))))((.((.(((((.....))))).)))).... ( -25.00, z-score = -3.55, R) >droVir3.scaffold_13047 702933 54 - 19223366 AGCCAAUACUCUACUCGUAUUGGCGUAGCUCGUGCUCCUGGCGUGUUCGGAUUG .((((((((.......))))))))(.(((.(((.......))).))))...... ( -15.00, z-score = -1.07, R) >droMoj3.scaffold_6540 4693103 54 - 34148556 AGCCGAUACUCUACUCGUAUUGGCGCAGUUCGUGCUCCUGGCGAGUGCGCAUUG .((((((((.......)))))))).((((.((..(((.....)))..)).)))) ( -21.70, z-score = -2.50, R) >droGri2.scaffold_14906 8552071 54 + 14172833 AGCCAAUACUCUAUUCGUAUUGGCGCAGCUCGUGCUCCUGGCGCGUUCGAAUUG .((((((((.......))))))))...(..(((((.....)))))..)...... ( -19.20, z-score = -2.68, R) >anoGam1.chr2R 29095411 54 - 62725911 AGCCCAUCCUGUACUCGUACUGGCGCAGUUCCUGCUCCUGGCGCGUUCGCAUUG .(((......(((....))).((.((((...)))).)).)))((....)).... ( -13.60, z-score = -0.37, R) >apiMel3.Group5 5502254 54 + 13386189 AGCCAAUACUCUACUCCUAUUGGCGGAGUUCUUGUUCGUGGAGAGUGCGAAUUG .(((((((.........))))))).........((((((.......)))))).. ( -12.00, z-score = -0.24, R) >triCas2.ChLG7 8412977 54 + 17478683 AGCCUCUUUUGUAUUCAUAUUGGCGCAGUUCGUGCUCGUGGAGGGUCCGGAUAG .(((.....((....))....)))...(((((..(((.....)))..))))).. ( -13.60, z-score = -0.40, R) >consensus AGCCAAUACUCUACUCGUAUUGGCGCAGCUCGUGCUCCUGGCGCGUGCGCAUUG .((((((((.......))))))))...(..(((((.....)))))..)...... (-14.58 = -14.96 + 0.38)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:00:56 2011