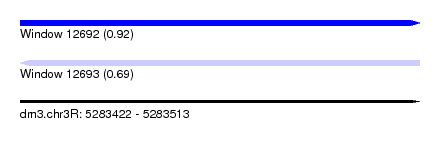
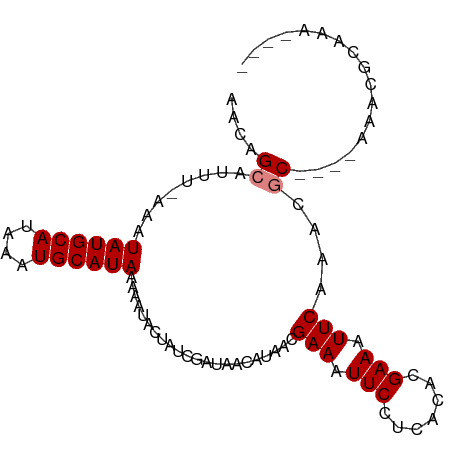
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 5,283,422 – 5,283,513 |
| Length | 91 |
| Max. P | 0.922655 |

| Location | 5,283,422 – 5,283,513 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 87.59 |
| Shannon entropy | 0.23114 |
| G+C content | 0.28474 |
| Mean single sequence MFE | -20.06 |
| Consensus MFE | -16.43 |
| Energy contribution | -16.43 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.922655 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 5283422 91 + 27905053 UGCGUUUGCGUUU----GCGUUUGAAUUUCGUGUGAGGAAUUUCGUUAUGUUAUCGAUAGUAUUUUUAUGCAUUUAUGCAUAUUU-AAAUGCUGUU ..((...((((..----(((...(((((((......)))))))))).))))...))(((((((((.((((((....))))))...-))))))))). ( -22.90, z-score = -2.05, R) >droSim1.chr3R 11466932 87 - 27517382 ----UUUGCGUUU----GCGUUUGAAUUUCGUGUGAGGAAUUUCGUUAUGUUAUCGAUAGUAUUUUUAUGCAUUUAUGCAUAUUU-AAAUGCUGUU ----...((((..----(((...(((((((......)))))))))).))))....((((((((((.((((((....))))))...-)))))))))) ( -21.40, z-score = -2.21, R) >droSec1.super_0 4507780 87 - 21120651 ----UUUGCGUUU----GCGUUUGAAUUUCGUGUGAGGAAUUUCGUUAUGUUAUCGAUAGUAUUUUUAUGCAUUUAUGCAUAUUU-AAAUGCUGUU ----...((((..----(((...(((((((......)))))))))).))))....((((((((((.((((((....))))))...-)))))))))) ( -21.40, z-score = -2.21, R) >droYak2.chr3R 9331895 81 + 28832112 ----------UUU----GCAUUUGAAUUUCGUGUGAGGAAUUUCGUUAUGUUAUCGAUAGUAUUUUUAUGCAUUUAUGCAUAUUU-AAAUGCUGUU ----------...----((((..(((((((......)))))))....))))....((((((((((.((((((....))))))...-)))))))))) ( -19.60, z-score = -2.55, R) >droEre2.scaffold_4770 1425834 81 - 17746568 ----------UUU----GCAUUUGAAUUUCGUGUGAGGAAUUUCGUUACGUUAUCGAUAGUAUUUUUAUGCAUUUAUGCAUAUUU-AAAUGCUGUU ----------...----(((((((((..((((((((.(.....).)))))....))).........((((((....)))))))))-)))))).... ( -18.00, z-score = -1.78, R) >dp4.chr2 27660058 94 - 30794189 -UUUUUGGAAUUUUAAAGUUUUUGAAUUUCGUGCU-GGAAUUUCGUUAUGUUAUCGAUAGUAUUUUUAUGCAUUUAUGCAUAUUUCAAAUGCUGUU -.....(((((((...(((...((.....)).)))-.)))))))...........((((((((((.((((((....))))))....)))))))))) ( -18.20, z-score = -1.65, R) >droPer1.super_7 1208313 94 + 4445127 -UUUUUGGAAUUUUGAAGUUUUUGAAUUUCGUGCU-GGAAUUUCGUUAUGUUAUCGAUAGUAUUUUUAUGCAUUUAUGCAUAUUUCAAAUGCUGUU -.....((((((((((((((....)))))))....-.)))))))...........((((((((((.((((((....))))))....)))))))))) ( -18.90, z-score = -1.66, R) >consensus ____UUUGCGUUU____GCGUUUGAAUUUCGUGUGAGGAAUUUCGUUAUGUUAUCGAUAGUAUUUUUAUGCAUUUAUGCAUAUUU_AAAUGCUGUU .......................(((((((......)))))))............((((((((((.((((((....))))))....)))))))))) (-16.43 = -16.43 + -0.00)

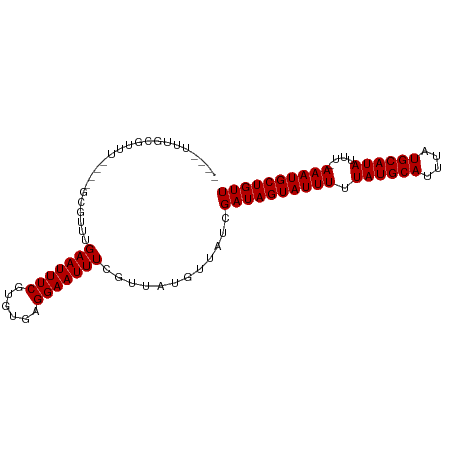
| Location | 5,283,422 – 5,283,513 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 87.59 |
| Shannon entropy | 0.23114 |
| G+C content | 0.28474 |
| Mean single sequence MFE | -11.19 |
| Consensus MFE | -7.20 |
| Energy contribution | -7.20 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.687541 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 5283422 91 - 27905053 AACAGCAUUU-AAAUAUGCAUAAAUGCAUAAAAAUACUAUCGAUAACAUAACGAAAUUCCUCACACGAAAUUCAAACGC----AAACGCAAACGCA ....((....-...((((((....))))))......................(((.(((.......))).)))....))----....((....)). ( -11.20, z-score = -1.73, R) >droSim1.chr3R 11466932 87 + 27517382 AACAGCAUUU-AAAUAUGCAUAAAUGCAUAAAAAUACUAUCGAUAACAUAACGAAAUUCCUCACACGAAAUUCAAACGC----AAACGCAAA---- ....((....-...((((((....))))))......................(((.(((.......))).)))....))----.........---- ( -9.90, z-score = -1.56, R) >droSec1.super_0 4507780 87 + 21120651 AACAGCAUUU-AAAUAUGCAUAAAUGCAUAAAAAUACUAUCGAUAACAUAACGAAAUUCCUCACACGAAAUUCAAACGC----AAACGCAAA---- ....((....-...((((((....))))))......................(((.(((.......))).)))....))----.........---- ( -9.90, z-score = -1.56, R) >droYak2.chr3R 9331895 81 - 28832112 AACAGCAUUU-AAAUAUGCAUAAAUGCAUAAAAAUACUAUCGAUAACAUAACGAAAUUCCUCACACGAAAUUCAAAUGC----AAA---------- ....((((((-...((((((....))))))......................(((.(((.......))).)))))))))----...---------- ( -13.40, z-score = -3.82, R) >droEre2.scaffold_4770 1425834 81 + 17746568 AACAGCAUUU-AAAUAUGCAUAAAUGCAUAAAAAUACUAUCGAUAACGUAACGAAAUUCCUCACACGAAAUUCAAAUGC----AAA---------- ....((((((-...((((((....))))))......................(((.(((.......))).)))))))))----...---------- ( -13.40, z-score = -3.03, R) >dp4.chr2 27660058 94 + 30794189 AACAGCAUUUGAAAUAUGCAUAAAUGCAUAAAAAUACUAUCGAUAACAUAACGAAAUUCC-AGCACGAAAUUCAAAAACUUUAAAAUUCCAAAAA- .......((((((.((((((....)))))).........(((.........)))......-.........))))))...................- ( -9.20, z-score = -1.54, R) >droPer1.super_7 1208313 94 - 4445127 AACAGCAUUUGAAAUAUGCAUAAAUGCAUAAAAAUACUAUCGAUAACAUAACGAAAUUCC-AGCACGAAAUUCAAAAACUUCAAAAUUCCAAAAA- .......((((((.((((((....))))))......................(((.(((.-.....))).)))......))))))..........- ( -11.30, z-score = -2.52, R) >consensus AACAGCAUUU_AAAUAUGCAUAAAUGCAUAAAAAUACUAUCGAUAACAUAACGAAAUUCCUCACACGAAAUUCAAACGC____AAACGCAAA____ ..............((((((....))))))......................(((.(((.......))).)))....................... ( -7.20 = -7.20 + 0.00)

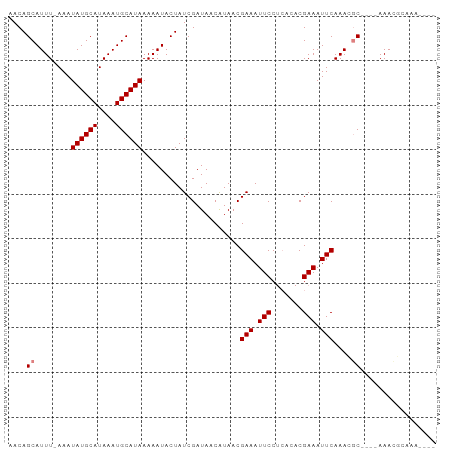
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:00:55 2011