| Sequence ID | dm3.chr3R |
|---|---|
| Location | 5,271,517 – 5,271,608 |
| Length | 91 |
| Max. P | 0.500000 |

| Location | 5,271,517 – 5,271,608 |
|---|---|
| Length | 91 |
| Sequences | 13 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 76.05 |
| Shannon entropy | 0.55490 |
| G+C content | 0.57785 |
| Mean single sequence MFE | -28.03 |
| Consensus MFE | -10.50 |
| Energy contribution | -10.68 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.61 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
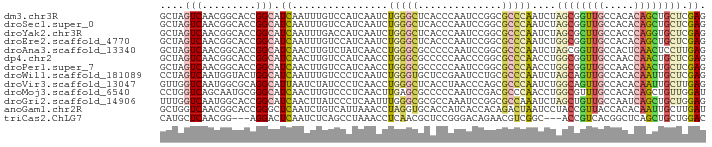
>dm3.chr3R 5271517 91 + 27905053 GCUAGUCAACGGCACCGGCAUCAAUUUGUCCAUCAAUCUGGGCUCACCCAAUCCGGCGCCCAAUCUAGCGGUUGCCACACAGCUGCUCGAG ..........(((.((((.........(((((......))))).........)))).)))...((.((((((((.....)))))))).)). ( -30.67, z-score = -2.77, R) >droSec1.super_0 4496122 91 - 21120651 GCUAGUCAACGGCACCGGCAUCAAUUUGUCCAUCAAUCUGGGCUCACCCAAUCCGGCGCCCAAUCUAGCGGUUGCCACACAGCUGCUCGAG ..........(((.((((.........(((((......))))).........)))).)))...((.((((((((.....)))))))).)). ( -30.67, z-score = -2.77, R) >droYak2.chr3R 9319310 91 + 28832112 GCUAGUCAACGGCACCGGCAUCAAUUUGACCAUCAAUCUGGGCUCACCCAAUCCGGCGCCCAAUCUAGCGCUUGCCACCCAGGUGCUCGAG (((((.....(((.((((.......(((.....)))..((((....))))..)))).)))....))))).((((.(((....)))..)))) ( -29.20, z-score = -1.97, R) >droEre2.scaffold_4770 1414214 91 - 17746568 GCUAGUCAACGGCACCGGCAUCAAUUUGUCCAUCAAUCUGGGCUCACCCAAUCCGGCGCCCAAUCUGGCGGUUGCCACACAGCUGCUCGAG ..........(((.((((.........(((((......))))).........)))).)))...((.((((((((.....)))))))).)). ( -30.67, z-score = -2.00, R) >droAna3.scaffold_13340 18920334 91 - 23697760 GCUAGUCAACGGCACCGGCAUCAACUUGUCUAUCAACCUGGGCGCCCCCAAUCCGGCGCCCAAUCUAGCGGUUGCCACUCAACUCCUUGAG ..........((((((((((......))))........((((((((........)))))))).......)).)))).(((((....))))) ( -32.70, z-score = -3.87, R) >dp4.chr2 27647844 91 - 30794189 GCUAGUCAACGGCACCGGCAUCAACUUGUCCAUCAACCUGGGCGCCCCCAACCCGGCGCCCAACCUGGCGGUUGCCAACCAACUGCUCGAG ((.(((....(((((((.((.....(((.....)))..((((((((........))))))))...)).))).)))).....)))))..... ( -32.20, z-score = -1.87, R) >droPer1.super_7 1195995 91 + 4445127 GCUAGUCAACGGCACCGGCAUCAACUUGUCCAUCAACCUGGGCGCCCCCAAUCCGGCGCCCAACCUGGCGGUUGCCAACCAACUGCUCGAG ((.(((....(((((((.((.....(((.....)))..((((((((........))))))))...)).))).)))).....)))))..... ( -32.20, z-score = -2.02, R) >droWil1.scaffold_181089 2642672 91 - 12369635 CCUAGUCAAUGGUACUGGCAUCAAUUUGUCCCUCAAUCUGGGUGCUCCGAAUCCUGCGCCCAAUCUAGCAGUUGCCACACAAUUGCUCGAG ((........))....((((......)))).(((....(((((((..........)))))))....((((((((.....)))))))).))) ( -24.90, z-score = -1.78, R) >droVir3.scaffold_13047 690616 91 + 19223366 GUUGGUCAAUGGCGCAGGCAUUAAUCUAUCCCUCAACCUGGGCUCACCUAACCCAGCGCCCAAUCUGGCAGUUGCCACACAAUUGCUUGAG ..........((((((((.((......)).))).....((((.........)))))))))...((.((((((((.....)))))))).)). ( -25.10, z-score = -0.59, R) >droMoj3.scaffold_6540 4680066 91 + 34148556 CCUGGUCAGCAAUGCGGGCAUCAACUUGUCCCUCAACUUGAGCGCCCCCAAUCCGACGCCCAACCUGGCGUUUGCCACACAGCUGUUGGAU .(..(.((((..((.((((.((((.(((.....))).))))..)))).))....((((((......)))))).........)))))..).. ( -29.80, z-score = -1.40, R) >droGri2.scaffold_14906 8537052 91 - 14172833 UUUGGUCAAUGGCACCGGCAUCAACUUAUCCCUCAAUUUGGGCGCGCCAAAUCCGGCGCCAAAUCUAGCUGUUGCCAAUCAGCUGCUGGAG .........((((.((((...........(((.......)))..........)))).))))..((((((.((((.....)))).)))))). ( -29.00, z-score = -1.03, R) >anoGam1.chr2R 33722691 91 - 62725911 GCUGGUCAACGGCACCGGGCUCAAUCUGUCAUUAAACCUAGGUGCACCAUCACCACAGACUAAUCCUACCGUUACCACACAAUUGCUUGAU ..((((.(((((....(((.....(((((...........((((......))))))))).....))).))))))))).............. ( -18.20, z-score = -0.40, R) >triCas2.ChLG7 8407849 85 - 17478683 CAUGCUCAACGG---AGGACUCAAUCUCAGCCUAAACCUCAACGCUCCGGGACAGAACGUCGGC---ACCGUCACGGCUCAGCUGCUGGAC ..((((...(((---((...........................))))).(((.....))))))---)..(((.((((......))))))) ( -19.13, z-score = 1.06, R) >consensus GCUAGUCAACGGCACCGGCAUCAAUUUGUCCAUCAACCUGGGCGCACCCAAUCCGGCGCCCAAUCUAGCGGUUGCCACACAGCUGCUCGAG ....(((.........))).((................(((((..............)))))....((((((((.....)))))))).)). (-10.50 = -10.68 + 0.18)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:00:52 2011