
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 5,269,469 – 5,269,541 |
| Length | 72 |
| Max. P | 0.634453 |

| Location | 5,269,469 – 5,269,541 |
|---|---|
| Length | 72 |
| Sequences | 11 |
| Columns | 78 |
| Reading direction | forward |
| Mean pairwise identity | 75.79 |
| Shannon entropy | 0.47336 |
| G+C content | 0.39055 |
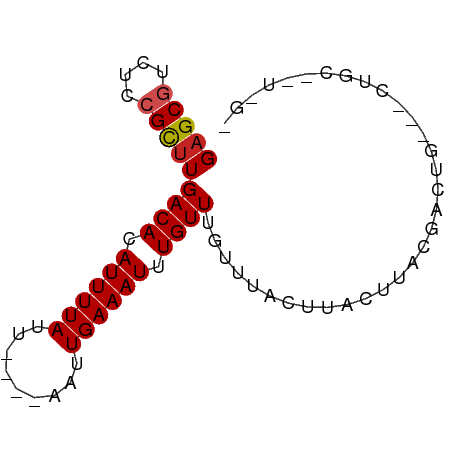
| Mean single sequence MFE | -12.06 |
| Consensus MFE | -7.06 |
| Energy contribution | -7.25 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.634453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
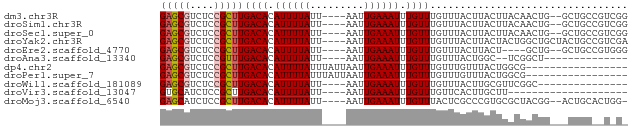
>dm3.chr3R 5269469 72 + 27905053 GAGCGUCUCCGCUUGACACAUUUUAUU----AAUUGAAAUUUGUUUGUUUACUUACUUACAACUG--GCUGCCGUCGG (((((....)))))((((.((((((..----...)))))).)))).................(((--((....))))) ( -13.00, z-score = -1.12, R) >droSim1.chr3R 11453229 72 - 27517382 GAGCGUCUCCGCUUGACACAUUUUAUU----AAUUGAAAUUUGUUUGUUUACUUACUUACAACUG--GCUGCCGUCGG (((((....)))))((((.((((((..----...)))))).)))).................(((--((....))))) ( -13.00, z-score = -1.12, R) >droSec1.super_0 4494082 72 - 21120651 GAGCGUCUCCGCUUGACACAUUUUAUU----AAUUGAAAUUUGUUUGUUUACUUACUUACAACUG--GCUGCCGUCGG (((((....)))))((((.((((((..----...)))))).)))).................(((--((....))))) ( -13.00, z-score = -1.12, R) >droYak2.chr3R 9317183 74 + 28832112 GAGCGUCUCCGCUUGACACAUUUUAUU----AAUUGAAAUUUGUUUGUUUACUUACUACUGGCUGCUACUGCCGUCGA (((((....)))))((((.((((((..----...)))))).))))..............((((.((....)).)))). ( -14.40, z-score = -1.63, R) >droEre2.scaffold_4770 1412106 68 - 17746568 GAGCGUCUCCGCUUGACACAUUUUAUU----AAUUGAAAUUUGUUUGUUUACUUACU----GCUG--GCUGCCGUGGG (((((....)))))((((.((((((..----...)))))).))))......(((((.----((..--...)).))))) ( -14.90, z-score = -1.29, R) >droAna3.scaffold_13340 18918188 58 - 23697760 GAGCGUCUCCGUUUGACACAUUUUAUU----AAUUGAAAUUUGUUUGUUUACUGGC--UCGGCU-------------- ((((..........((((.((((((..----...)))))).)))).........))--))....-------------- ( -10.51, z-score = -0.91, R) >dp4.chr2 27645462 61 - 30794189 GAGCGUCUCCGCUUGACACAUUUUAUUUAUUAAUUGAAAUUUGUUUGUUUGUUUACUGGCG----------------- (((((....)))))((((.((((((.........)))))).)))).((..(....)..)).----------------- ( -8.70, z-score = -0.55, R) >droPer1.super_7 1193606 61 + 4445127 GAGCGUCUCCGCUUGACACAUUUUAUUUAUUAAUUGAAAUUUGUUUGUUUGUUUACUGGCG----------------- (((((....)))))((((.((((((.........)))))).)))).((..(....)..)).----------------- ( -8.70, z-score = -0.55, R) >droWil1.scaffold_181089 2640367 59 - 12369635 GAGCGUCUCCGCUUGACACAUUUUAUU----AAUUGAAAUUUGUUUGUUUACUUGCGUUCGGC--------------- ((((((........((((.((((((..----...)))))).)))).........))))))...--------------- ( -12.43, z-score = -1.58, R) >droVir3.scaffold_13047 688388 54 + 19223366 GUGCAUCUCCGCUUGACACAUUUUAUU----AAUUGAAAUUUGUUUGUUCACUUGCUU-------------------- ..(((.....((..((((.((((((..----...)))))).)))).)).....)))..-------------------- ( -8.60, z-score = -1.57, R) >droMoj3.scaffold_6540 4678102 71 + 34148556 GAGCAUCUCCGCUUGACACAUUUUAUU----AAUUGAAAUUUGUUUACUCGCCCGUGCGCUACGG--ACUGCACUGG- ((((......))))((((.((((((..----...)))))).))))......((.((((.(....)--...)))).))- ( -15.40, z-score = -1.52, R) >consensus GAGCGUCUCCGCUUGACACAUUUUAUU____AAUUGAAAUUUGUUUGUUUACUUACUUACGACUG___CUGC__U_G_ (((((....)))))((((.((((((.........)))))).))))................................. ( -7.06 = -7.25 + 0.19)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:00:51 2011