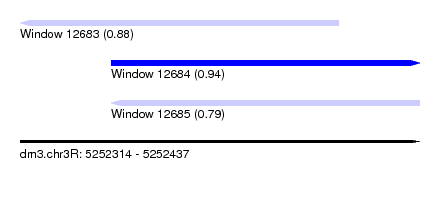
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 5,252,314 – 5,252,437 |
| Length | 123 |
| Max. P | 0.939984 |

| Location | 5,252,314 – 5,252,412 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 77.33 |
| Shannon entropy | 0.38530 |
| G+C content | 0.62208 |
| Mean single sequence MFE | -40.10 |
| Consensus MFE | -26.28 |
| Energy contribution | -27.32 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.878286 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
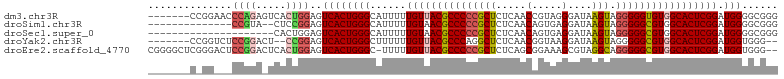
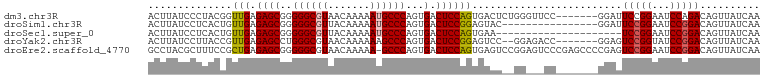
>dm3.chr3R 5252314 98 - 27905053 -------CCGGAACCCAGAGUCACUGGAGUCACUGGGCAUUUUUGUUACGCCCCCGCUCUCAACCGUAGGGAUAAGUAGGGGGUGUGGCACUCGGAUGGGGCGGG -------(((...((((((.((....)).)).(((((......((((((((((((...((...((....))...))..))))))))))))))))).)))).))). ( -40.40, z-score = -1.17, R) >droSim1.chr3R 11441529 89 + 27517382 --------------CCGUA--CUCCGGAGUCACUGGGCAUUUUUGUAACGCCCCCGCUCUCAACAGUGAGGAUAAGUAGGGGGCGUGGCACUCGGAUGGGGCGGG --------------((((.--((((((.(((((...(((....)))...(((((((((((((....))))....))).))))))))))).).)))).)..)))). ( -37.70, z-score = -1.78, R) >droSec1.super_0 4477233 84 + 21120651 ---------------------CACUGGAGUCACUGGGCAUUUUUGUAACGCCCCCGCUCUCAACAGUGAGGAUAAGUAGGGGGCGUGGCACUCGGAUGGGGCGGG ---------------------..((.(..((((((((......(((.(((((((((((((((....))))....))).)))))))).)))))))).)))..).)) ( -31.30, z-score = -1.08, R) >droYak2.chr3R 9299535 94 - 28832112 -------CCGGUCUCCGGACU--CCGGAGUCACUGGGCUUUUUUGUUACGCCCAGGCUCUCAACGGUAAGGAUAAGUAGGGGGCGUGGCACUCGGAUGGUGGG-- -------(((((((((((...--))))))..)))))(((.(((((((((((((..(((.....(.....)....)))...))))))))))...))).)))...-- ( -34.50, z-score = -0.37, R) >droEre2.scaffold_4770 1395248 102 + 17746568 CGGGGCUCGGGACUCCGGACUCACUGGAGUCACUGGGC-UUUUUGUUACGCCCCCGCUCUCAGCGGAAAGCGUAGGCAGGGGGCGUGGCACUCGGAUGGUGGG-- ((((((((((((((((((.....)))))))).))))))-....(((((((((((((((..(.((.....)))..))).)))))))))))))))).........-- ( -56.60, z-score = -4.41, R) >consensus _______C_GG___CCGGA_UCACUGGAGUCACUGGGCAUUUUUGUUACGCCCCCGCUCUCAACAGUAAGGAUAAGUAGGGGGCGUGGCACUCGGAUGGGGCGGG ..............((((.....))))..((((((((......(((((((((((((((.....(.....)....))).))))))))))))))))).)))...... (-26.28 = -27.32 + 1.04)
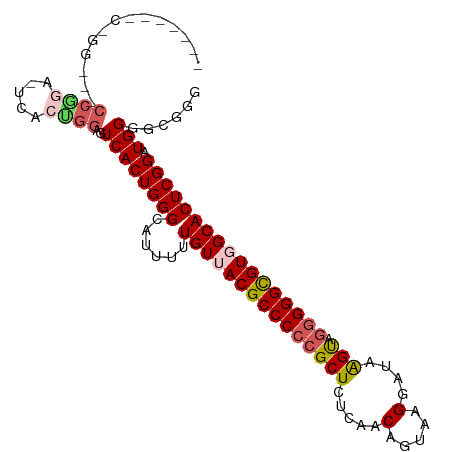
| Location | 5,252,342 – 5,252,437 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 77.51 |
| Shannon entropy | 0.38151 |
| G+C content | 0.53257 |
| Mean single sequence MFE | -34.58 |
| Consensus MFE | -16.06 |
| Energy contribution | -16.26 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.939984 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
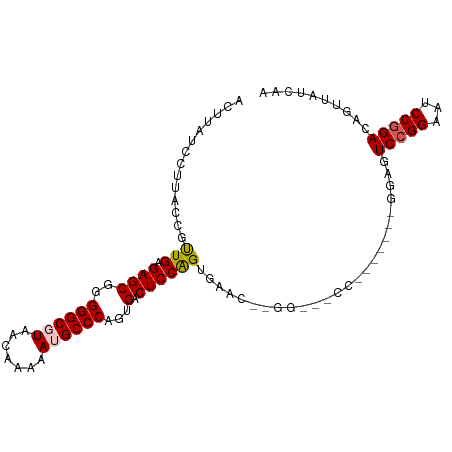
>dm3.chr3R 5252342 95 + 27905053 ACUUAUCCCUACGGUUGAGAGCGGGGGCGUAACAAAAAUGCCCAGUGACUCCAGUGACUCUGGGUUCC-------GGAUUCCGGAAUCCAGACAGUUAUCAA .((((.((....)).)))).((..((((((.......)))))).)).......(((((((((((((((-------((...))))))))))))..)))))... ( -35.50, z-score = -2.47, R) >droSim1.chr3R 11441557 86 - 27517382 ACUUAUCCUCACUGUUGAGAGCGGGGGCGUUACAAAAAUGCCCAGUGACUCCGGAGUAC----------------GGAUUCCGGAAUCCGGACAGUUAUCAA ..........((((((..((((..(((((((.....))))))).))...((((((((..----------------..)))))))).))..))))))...... ( -29.30, z-score = -1.84, R) >droSec1.super_0 4477261 81 - 21120651 ACUUAUCCUCACUGUUGAGAGCGGGGGCGUUACAAAAAUGCCCAGUGACUCCAGUGAA---------------------UCCGGAAUCCGGACAGUUAUCAA ........((((((..(((.((..(((((((.....))))))).))..))))))))).---------------------(((((...))))).......... ( -27.90, z-score = -2.54, R) >droYak2.chr3R 9299561 93 + 28832112 ACUUAUCCUUACCGUUGAGAGCCUGGGCGUAACAAAAAAGCCCAGUGACUCCGGAGUCC--GGAGACC-------GGAGUCCGGUAUCCGGACAGUUAUCAA ...........(((....((((((((((...........)))))).).)))))).((((--(((.(((-------((...))))).)))))))......... ( -38.80, z-score = -3.93, R) >droEre2.scaffold_4770 1395274 101 - 17746568 GCCUACGCUUUCCGCUGAGAGCGGGGGCGUAACAAAAA-GCCCAGUGACUCCAGUGAGUCCGGAGUCCCGAGCCCCGAGUCCGGAAUCCGGACAGUUAUCAA (((..(((((((....)))))))..)))(((((.....-...((.((....)).)).(((((((...(((..(.....)..)))..))))))).)))))... ( -41.40, z-score = -3.05, R) >consensus ACUUAUCCUUACCGUUGAGAGCGGGGGCGUAACAAAAAUGCCCAGUGACUCCAGUGAAC__GG___CC_______GGAGUCCGGAAUCCGGACAGUUAUCAA ..............(((.((((..((((((.......))))))...).)))))).........................(((((...))))).......... (-16.06 = -16.26 + 0.20)
| Location | 5,252,342 – 5,252,437 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 77.51 |
| Shannon entropy | 0.38151 |
| G+C content | 0.53257 |
| Mean single sequence MFE | -33.02 |
| Consensus MFE | -18.89 |
| Energy contribution | -18.73 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.793989 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
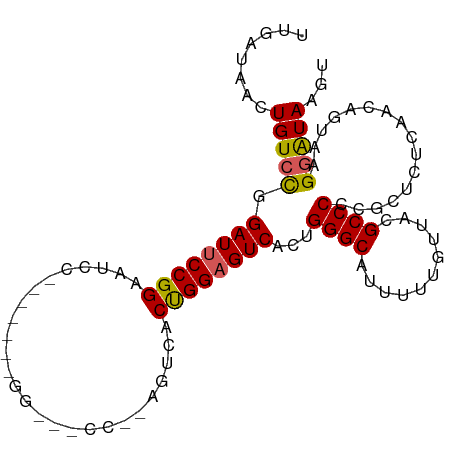
>dm3.chr3R 5252342 95 - 27905053 UUGAUAACUGUCUGGAUUCCGGAAUCC-------GGAACCCAGAGUCACUGGAGUCACUGGGCAUUUUUGUUACGCCCCCGCUCUCAACCGUAGGGAUAAGU .((((.....(((((.((((((...))-------)))).))))))))).((((((....((((...........))))..)))).)).((....))...... ( -27.60, z-score = -0.28, R) >droSim1.chr3R 11441557 86 + 27517382 UUGAUAACUGUCCGGAUUCCGGAAUCC----------------GUACUCCGGAGUCACUGGGCAUUUUUGUAACGCCCCCGCUCUCAACAGUGAGGAUAAGU ........(((((.(((((((((....----------------....)))))))))...((((.((.....)).)))).((((......)))).)))))... ( -28.10, z-score = -1.87, R) >droSec1.super_0 4477261 81 + 21120651 UUGAUAACUGUCCGGAUUCCGGA---------------------UUCACUGGAGUCACUGGGCAUUUUUGUAACGCCCCCGCUCUCAACAGUGAGGAUAAGU ........(((((.((((((((.---------------------....))))))))...((((.((.....)).)))).((((......)))).)))))... ( -24.10, z-score = -1.19, R) >droYak2.chr3R 9299561 93 - 28832112 UUGAUAACUGUCCGGAUACCGGACUCC-------GGUCUCC--GGACUCCGGAGUCACUGGGCUUUUUUGUUACGCCCAGGCUCUCAACGGUAAGGAUAAGU ........(((((...(((((((((((-------((((...--.))..)))))))).((((((...........))))))........))))).)))))... ( -38.50, z-score = -3.06, R) >droEre2.scaffold_4770 1395274 101 + 17746568 UUGAUAACUGUCCGGAUUCCGGACUCGGGGCUCGGGACUCCGGACUCACUGGAGUCACUGGGC-UUUUUGUUACGCCCCCGCUCUCAGCGGAAAGCGUAGGC .........((((((...))))))..(((((((((((((((((.....)))))))).))))))-)))...((((((..((((.....))))...)))))).. ( -46.80, z-score = -3.24, R) >consensus UUGAUAACUGUCCGGAUUCCGGAAUCC_______GG___CC__AGUCACUGGAGUCACUGGGCAUUUUUGUUACGCCCCCGCUCUCAACAGUAAGGAUAAGU ........(((((.((((((((..........................))))))))...((((...........))))................)))))... (-18.89 = -18.73 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:00:49 2011