| Sequence ID | dm3.chr3R |
|---|---|
| Location | 5,170,853 – 5,170,955 |
| Length | 102 |
| Max. P | 0.704675 |

| Location | 5,170,853 – 5,170,955 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 96.47 |
| Shannon entropy | 0.06150 |
| G+C content | 0.38779 |
| Mean single sequence MFE | -26.02 |
| Consensus MFE | -24.26 |
| Energy contribution | -24.86 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.665386 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
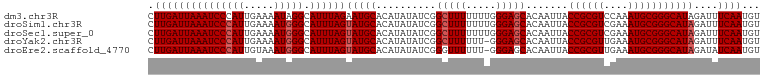
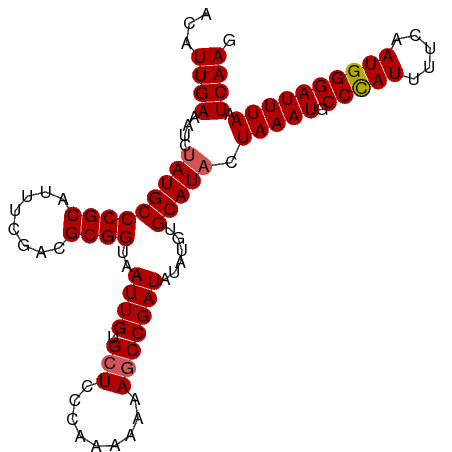
>dm3.chr3R 5170853 102 + 27905053 CUUGAUUAAAUCCCAUUGAAAAUAGGCAUUUAGAAUGCACAUAUAUCGGCUUUUUUUGGGAGCACAAUUACCGCGUCCAAAUGCGGGCAUAGAUUUCAAUGU .............((((((((....((((.....))))...(((..(.((....(((((..((.........))..))))).)).)..)))..)))))))). ( -23.10, z-score = -0.86, R) >droSim1.chr3R 11367693 102 - 27517382 CUUGAUUAAAUCCCAUUGAAAAUGGGCAUUUAGUAUGCACAUAUAUCGGCUUUUUUUGGGAGCACAAUUACCGCGUCGAAAUGCGGGCAUAGAUUUCAAUGU .(((((((((((((((.....))))).))))))(((((..........(((((.....))))).......((((((....)))))))))))....))))... ( -27.40, z-score = -1.80, R) >droSec1.super_0 4396279 102 - 21120651 CUUGAUUAAAUCCCAUUGAAAAUGGGCAUUUAGUAUGCACAUAUAUCGGCUUUUUUUGGGAGCACAAUUACCGCGUCGAAAUGCGGGCAUAGAUUUCAAUGU .(((((((((((((((.....))))).))))))(((((..........(((((.....))))).......((((((....)))))))))))....))))... ( -27.40, z-score = -1.80, R) >droYak2.chr3R 9217203 101 + 28832112 CUUGAUUAAAUCCCAUUGAAAAUGGGCAUUUAGUAUGCACAUAUAUCGGCUUUUUU-GGGAGCACAAUUACCGCGUUGAAAUGCGGGCAUAGAUUUCAAUGU .(((((((((((((((.....))))).))))))(((((..........(((((...-.))))).......((((((....)))))))))))....))))... ( -27.50, z-score = -1.85, R) >droEre2.scaffold_4770 1309960 101 - 17746568 CUUGAUUAAAUCCCAUUGUAAAUGGGCAUUUAGUAUGCACAUAUAUCGGGUUUUUU-GGGAGCACAAUUACCGCGUUGAAAUGCGGGCAUAGAUAUCAAUGU .(((((((((((((((.....))))).))))).(((((...........((((...-..)))).......((((((....)))))))))))...)))))... ( -24.70, z-score = -0.79, R) >consensus CUUGAUUAAAUCCCAUUGAAAAUGGGCAUUUAGUAUGCACAUAUAUCGGCUUUUUUUGGGAGCACAAUUACCGCGUCGAAAUGCGGGCAUAGAUUUCAAUGU .(((((((((((((((.....))))).))))))(((((..........(((((.....))))).......((((((....)))))))))))....))))... (-24.26 = -24.86 + 0.60)
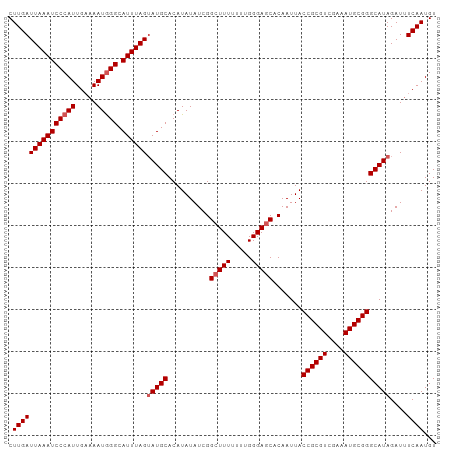
| Location | 5,170,853 – 5,170,955 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 96.47 |
| Shannon entropy | 0.06150 |
| G+C content | 0.38779 |
| Mean single sequence MFE | -22.54 |
| Consensus MFE | -20.68 |
| Energy contribution | -20.92 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.704675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
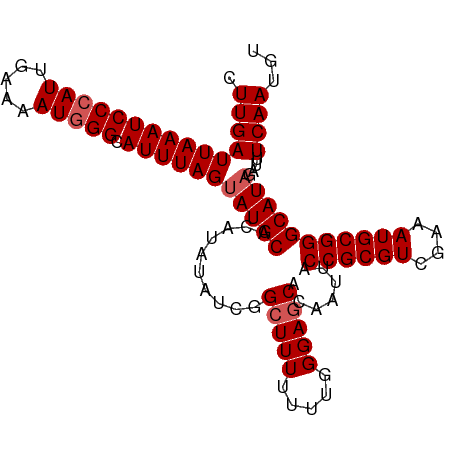
>dm3.chr3R 5170853 102 - 27905053 ACAUUGAAAUCUAUGCCCGCAUUUGGACGCGGUAAUUGUGCUCCCAAAAAAAGCCGAUAUAUGUGCAUUCUAAAUGCCUAUUUUCAAUGGGAUUUAAUCAAG .((((((((.........(((((((((((((...((((.(((.........)))))))...))))...)))))))))....))))))))............. ( -22.72, z-score = -1.12, R) >droSim1.chr3R 11367693 102 + 27517382 ACAUUGAAAUCUAUGCCCGCAUUUCGACGCGGUAAUUGUGCUCCCAAAAAAAGCCGAUAUAUGUGCAUACUAAAUGCCCAUUUUCAAUGGGAUUUAAUCAAG ...((((....(((((((((........))))..((((.(((.........)))))))......))))).(((((.(((((.....)))))))))).)))). ( -22.90, z-score = -1.58, R) >droSec1.super_0 4396279 102 + 21120651 ACAUUGAAAUCUAUGCCCGCAUUUCGACGCGGUAAUUGUGCUCCCAAAAAAAGCCGAUAUAUGUGCAUACUAAAUGCCCAUUUUCAAUGGGAUUUAAUCAAG ...((((....(((((((((........))))..((((.(((.........)))))))......))))).(((((.(((((.....)))))))))).)))). ( -22.90, z-score = -1.58, R) >droYak2.chr3R 9217203 101 - 28832112 ACAUUGAAAUCUAUGCCCGCAUUUCAACGCGGUAAUUGUGCUCCC-AAAAAAGCCGAUAUAUGUGCAUACUAAAUGCCCAUUUUCAAUGGGAUUUAAUCAAG ...((((....(((((((((........))))..((((.(((...-.....)))))))......))))).(((((.(((((.....)))))))))).)))). ( -23.70, z-score = -2.10, R) >droEre2.scaffold_4770 1309960 101 + 17746568 ACAUUGAUAUCUAUGCCCGCAUUUCAACGCGGUAAUUGUGCUCCC-AAAAAACCCGAUAUAUGUGCAUACUAAAUGCCCAUUUACAAUGGGAUUUAAUCAAG ...(((((...(((((..((((.....((.(((..(((......)-))...)))))....))))))))).(((((.(((((.....))))))))))))))). ( -20.50, z-score = -1.33, R) >consensus ACAUUGAAAUCUAUGCCCGCAUUUCGACGCGGUAAUUGUGCUCCCAAAAAAAGCCGAUAUAUGUGCAUACUAAAUGCCCAUUUUCAAUGGGAUUUAAUCAAG ...((((....(((((((((........))))..((((.(((.........)))))))......))))).(((((.(((((.....)))))))))).)))). (-20.68 = -20.92 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:00:40 2011