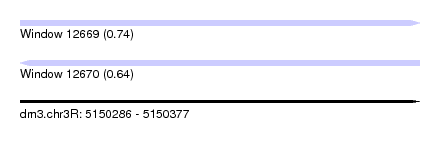
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 5,150,286 – 5,150,377 |
| Length | 91 |
| Max. P | 0.740225 |

| Location | 5,150,286 – 5,150,377 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 59.26 |
| Shannon entropy | 0.75661 |
| G+C content | 0.38529 |
| Mean single sequence MFE | -21.47 |
| Consensus MFE | -7.15 |
| Energy contribution | -8.16 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.740225 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
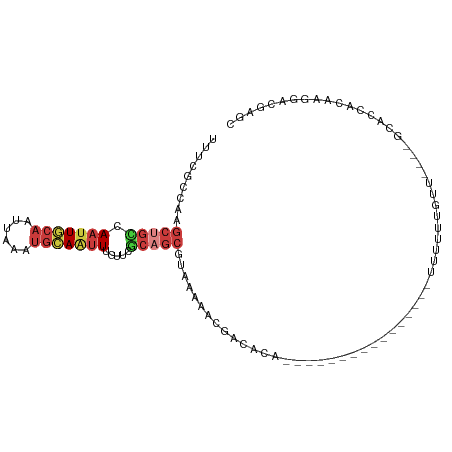
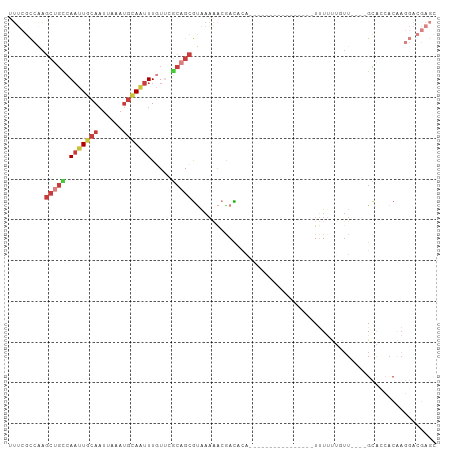
>dm3.chr3R 5150286 91 + 27905053 UUUCGCCAAGCUGUCAAUUGCAAUUAAAUGCAAUUUGUUCGCAGCGUAAAAACGACACAAUUU----------UUUUUUUUUUU----GCGCCACAAGGACGAGC .((((((..((...((((((((......))))).)))...)).(((((((((.((........----------...)).)))))----)))).....)).)))). ( -20.60, z-score = -0.84, R) >droPer1.super_7 150648 74 + 4445127 -----AUUUGCUCCCAAUUCCAAUUAAAUGCAGUUU-UCCCAAGCGUGCAUAAAAUAA-----------------UGUUCUCAUGAAUGGAAAACGA-------- -----............((((......(((((((((-....)))).))))).......-----------------.((((....)))))))).....-------- ( -11.80, z-score = -0.93, R) >droEre2.scaffold_4770 1289419 82 - 17746568 UUUCGCCGGGCUGCCAAUUGCAAUUAAAUGCAAUUUGUGCGCAGCGCAAAAACGACACA----------------UUU---GUU----GCACCACAAGGACGAGC .(((((((.(((((((((((((......))))))...)).))))).).....(((((..----------------..)---)))----)........)).)))). ( -24.90, z-score = -1.17, R) >droYak2.chr3R 9196502 85 + 28832112 UUUCGCCAAGCUGCCAAUUGCAAUUAAAUGCAAUUUGUUUGCUGCGUAAAAACGACACA----------------UUUUUUGUU----GCACCACGAGGACGAGU .((((((.(((.((.(((((((......))))))).))..))).(((.....(((((..----------------.....))))----)....))).)).)))). ( -20.30, z-score = -0.67, R) >droSec1.super_0 4375913 105 - 21120651 UUUCGCCAAGCUGCCAAUUGCAAUUAAAUGCAAUUUGUUCGCAGCGUAAAAACAACACAUUUUUUUGUUGUUUUUUUUUUUUUUGCUUGCACCACAAGGACGAGC .((((((..(((((.(((((((......))))))).....)))))..((((((((((........))))))))))......................)).)))). ( -26.90, z-score = -2.01, R) >droSim1.chr3R 11347087 102 - 27517382 UUUCGCCAAGCUGCCAAUUGCAAUUAAAUGCAAUUUGUUGGCAGCGUAAAAACGACAAAUUUUGUUGUUGUUUUUUUUUUUUUU---UGCAACACAAGGACGAGC .((((((..(((((((((((((......))))....)))))))))..(((((((((((......))))))))))).........---..........)).)))). ( -32.70, z-score = -3.30, R) >triCas2.ChLG6 5722945 75 - 13544221 GUACGUAAAAAUGUUAUUUUGACUUUUUCGGACUUUUUUUACUGAAAAAUUGAAAUGCC----------------UCUCCGGUUCCGGGUU-------------- ..............(((((..(.((((((((..........)))))))))..)))))..----------------..((((....))))..-------------- ( -13.10, z-score = -0.51, R) >consensus UUUCGCCAAGCUGCCAAUUGCAAUUAAAUGCAAUUUGUUCGCAGCGUAAAAACGACACA________________UUUUUUGUU____GCACCACAAGGACGAGC .........(((((.(((((((......))))))).....)))))............................................................ ( -7.15 = -8.16 + 1.00)

| Location | 5,150,286 – 5,150,377 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 59.26 |
| Shannon entropy | 0.75661 |
| G+C content | 0.38529 |
| Mean single sequence MFE | -21.87 |
| Consensus MFE | -7.66 |
| Energy contribution | -8.84 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.644749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
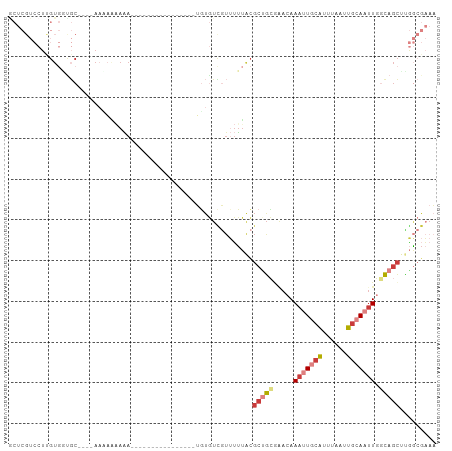
>dm3.chr3R 5150286 91 - 27905053 GCUCGUCCUUGUGGCGC----AAAAAAAAAAA----------AAAUUGUGUCGUUUUUACGCUGCGAACAAAUUGCAUUUAAUUGCAAUUGACAGCUUGGCGAAA ..(((((.....(((((----((.........----------...)))))))........((((......(((((((......)))))))..))))..))))).. ( -23.00, z-score = -0.97, R) >droPer1.super_7 150648 74 - 4445127 --------UCGUUUUCCAUUCAUGAGAACA-----------------UUAUUUUAUGCACGCUUGGGA-AAACUGCAUUUAAUUGGAAUUGGGAGCAAAU----- --------..(((((((...(((((((...-----------------...)))))))........)))-))))(((.((((((....)))))).)))...----- ( -13.90, z-score = -0.75, R) >droEre2.scaffold_4770 1289419 82 + 17746568 GCUCGUCCUUGUGGUGC----AAC---AAA----------------UGUGUCGUUUUUGCGCUGCGCACAAAUUGCAUUUAAUUGCAAUUGGCAGCCCGGCGAAA ..(((((...(..((((----((.---(((----------------((...)))))))))))..)((...(((((((......))))))).)).....))))).. ( -28.50, z-score = -1.40, R) >droYak2.chr3R 9196502 85 - 28832112 ACUCGUCCUCGUGGUGC----AACAAAAAA----------------UGUGUCGUUUUUACGCAGCAAACAAAUUGCAUUUAAUUGCAAUUGGCAGCUUGGCGAAA ..(((.((....(.(((----.........----------------(((..(((....)))..)))....(((((((......))))))).))).)..))))).. ( -19.90, z-score = -0.03, R) >droSec1.super_0 4375913 105 + 21120651 GCUCGUCCUUGUGGUGCAAGCAAAAAAAAAAAAAACAACAAAAAAAUGUGUUGUUUUUACGCUGCGAACAAAUUGCAUUUAAUUGCAAUUGGCAGCUUGGCGAAA ..(((((((((.....))))..........((((((((((........))))))))))..(((((.....(((((((......))))))).)))))..))))).. ( -30.80, z-score = -2.50, R) >droSim1.chr3R 11347087 102 + 27517382 GCUCGUCCUUGUGUUGCA---AAAAAAAAAAAAAACAACAACAAAAUUUGUCGUUUUUACGCUGCCAACAAAUUGCAUUUAAUUGCAAUUGGCAGCUUGGCGAAA ..(((((...........---.........((((((.((((......)))).))))))..(((((((....((((((......)))))))))))))..))))).. ( -29.20, z-score = -2.88, R) >triCas2.ChLG6 5722945 75 + 13544221 --------------AACCCGGAACCGGAGA----------------GGCAUUUCAAUUUUUCAGUAAAAAAAGUCCGAAAAAGUCAAAAUAACAUUUUUACGUAC --------------...(((....)))...----------------(((.((((.((((((.......))))))..))))..))).................... ( -7.80, z-score = 0.68, R) >consensus GCUCGUCCUUGUGGUGC____AAAAAAAAA________________UGUGUCGUUUUUACGCUGCGAACAAAUUGCAUUUAAUUGCAAUUGGCAGCUUGGCGAAA ............................................................(((((.....(((((((......))))))).)))))......... ( -7.66 = -8.84 + 1.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:00:36 2011