
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 5,149,045 – 5,149,165 |
| Length | 120 |
| Max. P | 0.981601 |

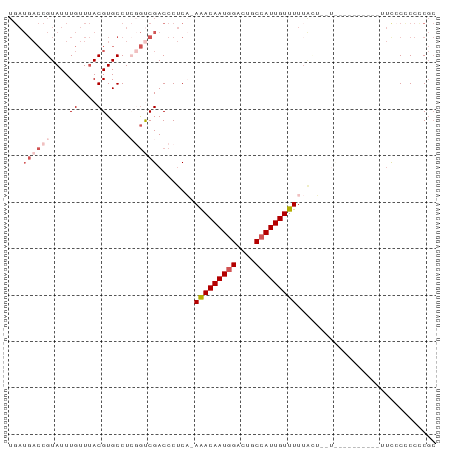
| Location | 5,149,045 – 5,149,136 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 71.56 |
| Shannon entropy | 0.50649 |
| G+C content | 0.50556 |
| Mean single sequence MFE | -16.42 |
| Consensus MFE | -8.92 |
| Energy contribution | -10.76 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.931551 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 5149045 91 + 27905053 UGAUGACCGUAUUUGUUUACGUGCCUCGGUCGACCCUCA-AAACAAUGGACUGCCAUUGUUUUUAGUUUUCCCACCCCGCUUCACCCCUCGC (((((((((....((....)).....))))))...((.(-(((((((((....)))))))))).))...............)))........ ( -19.40, z-score = -1.93, R) >droAna3.scaffold_13340 18801126 82 - 23697760 --------GUAUUUGUUAACGUGCCUGAACCGACCCCCCUAGACAAUAGACCUCCAUUGUUUUUCUGGUUUGCCAGCCCCUACCCCCACC-- --------............(((.................(((((((.(....).)))))))..((((....))))..........))).-- ( -9.50, z-score = -0.46, R) >droEre2.scaffold_4770 1288148 87 - 17746568 UGAUGACCGUAUUUGUUUACGUGCCUCGGCCGACCCUCC-AAACAAUGGACUGCCAUUGUUUUAGCUCCUCCAGUU----UUUCCUCCCCGC .((.(..((((......))))..).))((..((.(....-(((((((((....)))))))))..).))..))....----............ ( -15.40, z-score = -0.68, R) >droYak2.chr3R 9195171 77 + 28832112 UGAUGACCGUAUUUGUUUACGUGCCUGCGUCGACCCUCA-AAACAAUGGACUGCCAUUGUUUUUGCU--------------UUCCUCCCCGC .((.((..(((...(((.((((....)))).)))....(-(((((((((....))))))))))))).--------------.)).))..... ( -16.70, z-score = -2.26, R) >droSim1.chr3R 11345445 78 - 27517382 UGAUGACCGUAUUUGUUUACGUGCCUCGGUCGACCCUCA-AAACAAUGGACUGCCAUUGUUUUUAGU-------------UUCCCCCCUCGC .((.(..((((......))))..).))((..((..((.(-(((((((((....)))))))))).)).-------------.))..))..... ( -21.10, z-score = -3.36, R) >consensus UGAUGACCGUAUUUGUUUACGUGCCUCGGUCGACCCUCA_AAACAAUGGACUGCCAUUGUUUUUACU__U__________UUCCCCCCCCGC ...((((((....((....)).....))))))........(((((((((....))))))))).............................. ( -8.92 = -10.76 + 1.84)


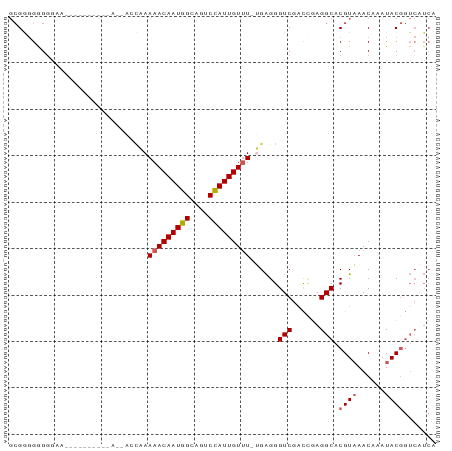
| Location | 5,149,045 – 5,149,136 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 71.56 |
| Shannon entropy | 0.50649 |
| G+C content | 0.50556 |
| Mean single sequence MFE | -24.16 |
| Consensus MFE | -11.42 |
| Energy contribution | -11.86 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.981601 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 5149045 91 - 27905053 GCGAGGGGUGAAGCGGGGUGGGAAAACUAAAAACAAUGGCAGUCCAUUGUUU-UGAGGGUCGACCGAGGCACGUAAACAAAUACGGUCAUCA ......(((((.((..(((.((....((.((((((((((....)))))))))-).))..)).)))...)).((((......)))).))))). ( -26.80, z-score = -2.03, R) >droAna3.scaffold_13340 18801126 82 + 23697760 --GGUGGGGGUAGGGGCUGGCAAACCAGAAAAACAAUGGAGGUCUAUUGUCUAGGGGGGUCGGUUCAGGCACGUUAACAAAUAC-------- --(.(((.(((..((((((((...((......(((((((....))))))).....)).))))))))..)).).))).)......-------- ( -23.10, z-score = -2.76, R) >droEre2.scaffold_4770 1288148 87 + 17746568 GCGGGGAGGAAA----AACUGGAGGAGCUAAAACAAUGGCAGUCCAUUGUUU-GGAGGGUCGGCCGAGGCACGUAAACAAAUACGGUCAUCA ............----....((..((.((.(((((((((....)))))))))-...)).))..))(((((.((((......))))))).)). ( -23.10, z-score = -1.16, R) >droYak2.chr3R 9195171 77 - 28832112 GCGGGGAGGAA--------------AGCAAAAACAAUGGCAGUCCAUUGUUU-UGAGGGUCGACGCAGGCACGUAAACAAAUACGGUCAUCA (((..((....--------------....((((((((((....)))))))))-).....))..))).(((.((((......))))))).... ( -21.72, z-score = -2.60, R) >droSim1.chr3R 11345445 78 + 27517382 GCGAGGGGGGAA-------------ACUAAAAACAAUGGCAGUCCAUUGUUU-UGAGGGUCGACCGAGGCACGUAAACAAAUACGGUCAUCA .....((..((.-------------.((.((((((((((....)))))))))-).))..))..))(((((.((((......))))))).)). ( -26.10, z-score = -3.62, R) >consensus GCGGGGGGGGAA__________A__ACCAAAAACAAUGGCAGUCCAUUGUUU_UGAGGGUCGACCGAGGCACGUAAACAAAUACGGUCAUCA ...........................((.(((((((((....))))))))).))...(((......))).((((......))))....... (-11.42 = -11.86 + 0.44)


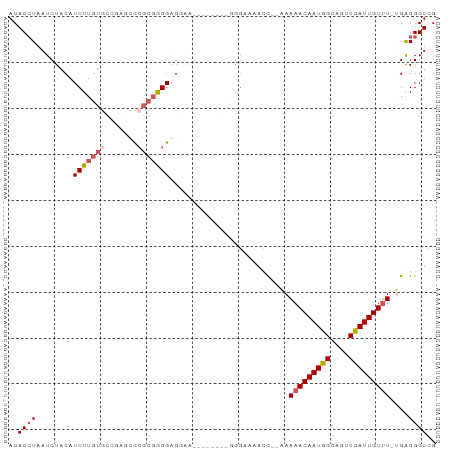
| Location | 5,149,075 – 5,149,165 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 70.59 |
| Shannon entropy | 0.52501 |
| G+C content | 0.49683 |
| Mean single sequence MFE | -24.48 |
| Consensus MFE | -11.98 |
| Energy contribution | -12.98 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.565333 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 5149075 90 - 27905053 ACACCUAAUCUACAUCUUGUGCCAUGCCCGCGAGGGGUGAAGCGGGGUGGGAAAACU--AAAAACAAUGGCAGUCCAUUGUUU-UGAGGGUCG ..(((...(((.((((((((..(((.(((....))))))..)))))))))))...((--.((((((((((....)))))))))-).))))).. ( -33.90, z-score = -2.68, R) >droAna3.scaffold_13340 18801148 89 + 23697760 ACACAUAAUCUACAUCUUGUACUCUCUUAGGUGGGGGUAGG----GGCUGGCAAACCAGAAAAACAAUGGAGGUCUAUUGUCUAGGGGGGUCG ..................(.((((((((((...........----..((((....))))....(((((((....)))))))))))))))))). ( -22.70, z-score = -0.45, R) >droEre2.scaffold_4770 1288178 86 + 17746568 ACACCUAAUCUACAUCUUGUGUGGAGCCCGCGGGGAGGAAA----AACUGGAGGAGC--UAAAACAAUGGCAGUCCAUUGUUU-GGAGGGUCG ..((((..(((((((...)))))))((((.(.((.......----..)).).)).))--..(((((((((....)))))))))-...)))).. ( -24.30, z-score = -0.30, R) >droYak2.chr3R 9195201 76 - 28832112 ACACCUAAUCUACAUCUUGUGCCGAGUCCGCGGGGAGGAA--------------AGC--AAAAACAAUGGCAGUCCAUUGUUU-UGAGGGUCG ..((((........((((.(.(((......))).))))).--------------...--.((((((((((....)))))))))-)..)))).. ( -19.00, z-score = -0.09, R) >droSim1.chr3R 11345475 69 + 27517382 ACACCUAAUCUACAUCUUG--------CCGCGAGG-------------GGGGAAACU--AAAAACAAUGGCAGUCCAUUGUUU-UGAGGGUCG ..(((.........((((.--------((....))-------------.))))..((--.((((((((((....)))))))))-).))))).. ( -22.50, z-score = -2.71, R) >consensus ACACCUAAUCUACAUCUUGUGCCGAGCCCGCGGGGAGGAA________GGGAAAACC__AAAAACAAUGGCAGUCCAUUGUUU_UGAGGGUCG ..((((........(((((((.......)))))))..........................(((((((((....)))))))))....)))).. (-11.98 = -12.98 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:00:34 2011