
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 5,144,502 – 5,144,593 |
| Length | 91 |
| Max. P | 0.741322 |

| Location | 5,144,502 – 5,144,593 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 56.64 |
| Shannon entropy | 0.87196 |
| G+C content | 0.42788 |
| Mean single sequence MFE | -23.84 |
| Consensus MFE | -8.08 |
| Energy contribution | -7.98 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.17 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.741322 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 5144502 91 - 27905053 AAAAAAAAAAAAACACAGGC---AAUGCGAACAC------AUCUAUGUGUAUCUCGUUGUAGCCACAGGCGUGUUACGCAUACGCCACGUAUGCAAAUCG ...........(((((..((---((((.((((((------(....))))).)).)))))).(((...))))))))..(((((((...)))))))...... ( -28.70, z-score = -3.81, R) >droSec1.super_0 4370576 89 + 21120651 --AAAACAAAAAAAACAGGC---AAUACGAACAC------AGCUAUGUGUAUCUCGUUGUAGCUACAGGCGUGUUACGCAUACGCCACGUAUGCAAAUCC --................((---(.((((.....------(((((((..........)))))))...(((((((.....))))))).)))))))...... ( -22.60, z-score = -1.71, R) >droYak2.chr3R 9190958 88 - 28832112 ---------AAAAAACAGGC---AGUGUGAACUUCUAUAUACAUAUAUGUAUCCCGUUGUAGCUACAGGCGUGUUACGCAUACGCCACGUAUGCAAAACG ---------....((((.((---..((((.((......(((((....)))))......))...)))).)).))))..(((((((...)))))))...... ( -19.40, z-score = -0.03, R) >droEre2.scaffold_4770 1283951 85 + 17746568 ----CAAAAAAAAUAUAGGC---GAUACGAACUGC--------CAUGUGUAUCUCGUUGUAGCUACAGGCGUGUUACGCAUACGCCACGUAUGCAAAUCG ----..............((---.(((((..((((--------.(((.......))).)))).....(((((((.....))))))).)))))))...... ( -21.30, z-score = -0.68, R) >droAna3.scaffold_13340 18797013 91 + 23697760 --------AAACUGGCUGGCAUACACACGUAUGUAUGUAUGUAUGGAUGUACCAUGUGCGUUAUUUGGACACGUUACGUAUACGCCGGGUAUGCAAAUC- --------...(((((..((((((....)))))).((((((((..(.(((.(((...........)))))))..)))))))).)))))...........- ( -24.40, z-score = 0.72, R) >dp4.chr2 26583727 96 + 30794189 ---CAGAGAUAUGUAAGAGUAUGUAAAUGACUGCAUACAUAUGCAUGUGUGUGGAAAGUGAUGUUUGGCCAUGUUGCGCAUACGCCGUGUAUGCAAAUU- ---((.(.(((((((...(((((((......)))))))...))))))).).))....(..((((......))))..)((((((.....)))))).....- ( -26.50, z-score = -0.78, R) >droPer1.super_7 144861 96 - 4445127 ---CAGAGAUAUGUAAGAGUAUGUAAAUGACUGCAUACAUAUGCAUGUGUGUGGAAAGUGAUGUUUGGCCAUGUUGCGCAUACGCCGUGUAUGCAAAUU- ---((.(.(((((((...(((((((......)))))))...))))))).).))....(..((((......))))..)((((((.....)))))).....- ( -26.50, z-score = -0.78, R) >droVir3.scaffold_12963 6959111 96 - 20206255 ----GCAAUAUAUCACAAGCACAAACACAAGCAAACGGACACACAGGCAUAUAAGUGUGUACAUCUGUCAAGGUUACGCAUACGCCACGUAUGCAAUGCG ----(((................(((....(((.....((((((..........)))))).....)))....)))..(((((((...)))))))..))). ( -21.30, z-score = -0.86, R) >consensus ____AAAAAAAAAUACAGGC___AAUACGAACACAU__AUAUACAUGUGUAUCUCGUUGUAGCUACAGGCAUGUUACGCAUACGCCACGUAUGCAAAUCG .............................................................................((((((.....))))))...... ( -8.08 = -7.98 + -0.11)

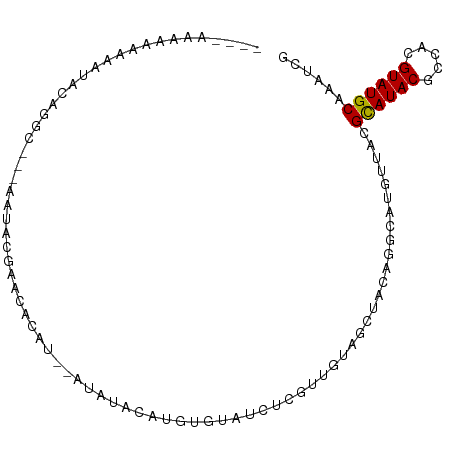

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:00:31 2011