| Sequence ID | dm3.chr3R |
|---|---|
| Location | 5,118,563 – 5,118,672 |
| Length | 109 |
| Max. P | 0.853070 |

| Location | 5,118,563 – 5,118,672 |
|---|---|
| Length | 109 |
| Sequences | 9 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.40 |
| Shannon entropy | 0.54304 |
| G+C content | 0.45172 |
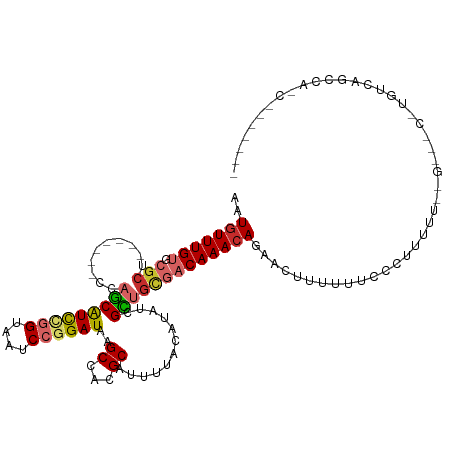
| Mean single sequence MFE | -22.45 |
| Consensus MFE | -13.15 |
| Energy contribution | -13.70 |
| Covariance contribution | 0.55 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.853070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
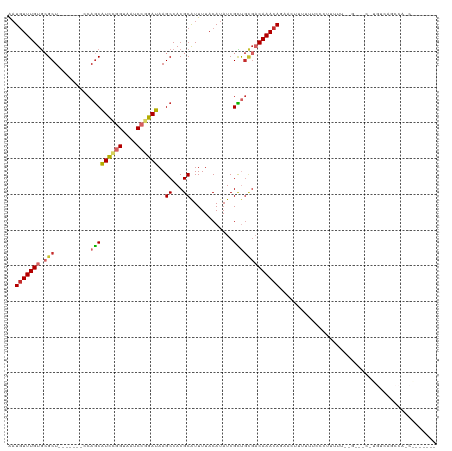
>dm3.chr3R 5118563 109 - 27905053 AAUGUUUGUGCGCUCCAGCAUCCAGCAUCCGGUAAUCCGGAUAAGCCACGCGUUUUACAUAUCGCUGCGACAAACACAACUUUUUUCCCUUUUC------CCUGUCAGCCAUCAG----- ..((((((((((((...((.....))((((((....)))))).)))...(((..........))).)).)))))))..................------.(((........)))----- ( -24.20, z-score = -2.20, R) >droWil1.scaffold_181089 9270928 90 + 12369635 GCUCUUUGUAAGCU--------CAGCAUUUGGUAAUCCGGAUAAGCCACGCAUUUUACAUAUCGUU--CACAAAAAUAACAUUUUAGUGGCAUGCUGCAG-------------------- (((.......))).--------((((((((((....))))))..(((((.................--..................)))))..))))...-------------------- ( -17.45, z-score = -0.38, R) >droPer1.super_7 124305 89 - 4445127 AAUGUUUGUGCGCU--------CAGCGUUGAGUAAUCCGGAUAAGCCACGCAUUUUACAUAUCGUCGUGUCAAACAGAAAAAGUGGUCGGGUAUCGG----------------------- ...........(((--------(......)))).(((((.....(((((...((((((((......)))).....))))...)))))))))).....----------------------- ( -15.40, z-score = 1.72, R) >dp4.chr2 26564272 89 + 30794189 AAUGUUUGUGCGCU--------CAGCGUUGAGUAAUCCGGAUAAGCCACGCAUUUUACAUAUCGUCGUGUCAAACAGAAAAAGUGGUCGGGUAUCGG----------------------- ...........(((--------(......)))).(((((.....(((((...((((((((......)))).....))))...)))))))))).....----------------------- ( -15.40, z-score = 1.72, R) >droAna3.scaffold_13340 18778050 91 + 23697760 AAUGUUUGUGGGCU-------GCAACAUUCGGUAAUCCGGAUAAGCCACGCAUUUUACAUAUCGUUGUGACAAACAUAAAAUGUUUUUCUGCCAUCGA---------------------- .((((.((((((((-------.....((((((....)))))).)))).))))....)))).(((.((.((.((((((...)))))).))...)).)))---------------------- ( -23.50, z-score = -1.53, R) >droEre2.scaffold_4770 1264864 105 + 17746568 AAUGUUUGUGCGCU-------CCAGCAUCCGGUAAUCCGGAUAAGCCACGCAUUUUACAUAUCGCUGCGACAAACAGAAGUUUUUUCCCUUUUUUUG---CUUGUCAGCCAACGG----- .((((..((((((.-------...))((((((....)))))).......))))...))))...(((((((((((.(((((........)))))))))---.))).))))......----- ( -29.00, z-score = -2.48, R) >droYak2.chr3R 9171822 113 - 28832112 AAUGUUUGUGCGCU-------CCAGCAUCCGGUAAUCCGGAUAAGCCACGCAUUUUACAUAUCGCUGCGACAAACAGAACUUUAUUCCCUUUUUUUGUCCCUUGUCAGCCAUCAGAAGGG ..(((((((.(((.-------..(((((((((....))))))..((...))............)))))))))))))..........(((((((..((.(........).))..))))))) ( -30.10, z-score = -2.50, R) >droSec1.super_0 4352072 109 + 21120651 AAUGUUUGUGCGCUCCAGCAUCCAGCAUCCGGUAAUCCGGAUAAGCCACGCAUUUUACAUAUCGCUGCGACAAACACAACUUUUUUCCCUUUUC------CCUGUCAGCCAUCAU----- ..(((((((((......)))((((((((((((....))))))..((...))............)))).))))))))..................------...............----- ( -23.30, z-score = -2.18, R) >droSim1.chr3R 11320800 109 + 27517382 AAUGUUUGUGCGCUCCAGCAUCCAGCAUCCGGUAAUCCGGAUAAGCCACGCAUUUUACAUAUCGCUGCGACAAACACAACUUUUUUCCCUUUUC------CCUGUCAGCCAUCAG----- ..(((((((((......)))((((((((((((....))))))..((...))............)))).))))))))..................------.(((........)))----- ( -23.70, z-score = -2.39, R) >consensus AAUGUUUGUGCGCU_______CCAGCAUCCGGUAAUCCGGAUAAGCCACGCAUUUUACAUAUCGCUGCGACAAACAGAACUUUUUUCCCUUUUU__G___C_UGUCAGCCA_C_______ ..(((((((.(((.............((((((....))))))..((...))...............))))))))))............................................ (-13.15 = -13.70 + 0.55)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:00:30 2011