
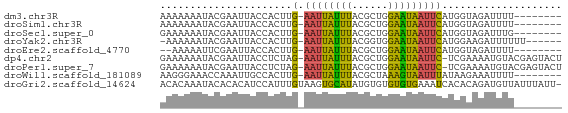
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 5,099,720 – 5,099,778 |
| Length | 58 |
| Max. P | 0.803623 |

| Location | 5,099,720 – 5,099,778 |
|---|---|
| Length | 58 |
| Sequences | 9 |
| Columns | 67 |
| Reading direction | forward |
| Mean pairwise identity | 71.36 |
| Shannon entropy | 0.58908 |
| G+C content | 0.28051 |
| Mean single sequence MFE | -12.29 |
| Consensus MFE | -2.45 |
| Energy contribution | -2.02 |
| Covariance contribution | -0.43 |
| Combinations/Pair | 1.75 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.20 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.803623 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 5099720 58 + 27905053 AAAAAAAUACGAAUUACCACUUG-AAUUAUUUACGCUGGAAUAAUUCAUGGUAGAUUUU-------- .............((((((..((-((((((((......)))))))))))))))).....-------- ( -13.50, z-score = -4.38, R) >droSim1.chr3R 11306809 58 - 27517382 AAAAAAAUACGAAUUACCACUUG-AAUUAUUUACGCUGGAAUAAUUCAUGGUAGAUUUU-------- .............((((((..((-((((((((......)))))))))))))))).....-------- ( -13.50, z-score = -4.38, R) >droSec1.super_0 4338635 58 - 21120651 GAAAAAAUACGAAUUACCACUUG-AAUUAUUUACGCUGGAAUAAUUCAUGGUAGAUUUG-------- .........((((((((((..((-((((((((......)))))))))))))).))))))-------- ( -14.70, z-score = -4.87, R) >droYak2.chr3R 9157840 59 + 28832112 -AAAAAAUACGAAUUACCACUUG-AAUUAUUUACGGUGGAAUAAUUCAUGGAAGAUUUUUU------ -...............(((..((-((((((((......)))))))))))))..........------ ( -10.40, z-score = -2.14, R) >droEre2.scaffold_4770 1250115 56 - 17746568 --AAAAAUUCGAAUUACCACUUG-AAUUAUUUACGCUGGAAUAAUUCAUGGUAGAUUUU-------- --...........((((((..((-((((((((......)))))))))))))))).....-------- ( -13.50, z-score = -3.97, R) >dp4.chr2 26559263 65 - 30794189 GAAAAAAUACGAAUUACCUCUAG-AAUUAUUUACGCUGGAAUAAUUC-UCGAAAAUGUACGAGUACU .........((.((....((.((-((((((((......)))))))))-).))....)).))...... ( -9.80, z-score = -1.70, R) >droPer1.super_7 119322 65 + 4445127 GAAAAAAUACGAAUUACCUCUAG-AAUUAUUUACGCUGGAAUAAUUC-UCGAAAAUGUACGAGUACU .........((.((....((.((-((((((((......)))))))))-).))....)).))...... ( -9.80, z-score = -1.70, R) >droWil1.scaffold_181089 9261871 58 - 12369635 AAGGGAAACCAAAUUGCCACUUG-AAUUAUUUACGCUAAAGUAAUUUAUAAGAAAUUUU-------- ..((....))(((((......((-((((((((......)))))))))).....))))).-------- ( -10.50, z-score = -2.45, R) >droGri2.scaffold_14624 3809815 66 + 4233967 ACACAAAUACACACAUCCAUUUGUAAGUGCAUAUGUGUGUGUGAAAUCACACAGAUGUUAUUUAUU- (((((.((.((((((......)))..))).)).)))))(((((....)))))..............- ( -14.90, z-score = -0.70, R) >consensus AAAAAAAUACGAAUUACCACUUG_AAUUAUUUACGCUGGAAUAAUUCAUGGUAGAUUUU________ ........................((((((((......))))))))..................... ( -2.45 = -2.02 + -0.43)

| Location | 5,099,720 – 5,099,778 |
|---|---|
| Length | 58 |
| Sequences | 9 |
| Columns | 67 |
| Reading direction | reverse |
| Mean pairwise identity | 71.36 |
| Shannon entropy | 0.58908 |
| G+C content | 0.28051 |
| Mean single sequence MFE | -10.45 |
| Consensus MFE | -3.07 |
| Energy contribution | -2.73 |
| Covariance contribution | -0.34 |
| Combinations/Pair | 1.62 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.669862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 5099720 58 - 27905053 --------AAAAUCUACCAUGAAUUAUUCCAGCGUAAAUAAUU-CAAGUGGUAAUUCGUAUUUUUUU --------......((((((((((((((........)))))))-))..))))).............. ( -10.80, z-score = -3.34, R) >droSim1.chr3R 11306809 58 + 27517382 --------AAAAUCUACCAUGAAUUAUUCCAGCGUAAAUAAUU-CAAGUGGUAAUUCGUAUUUUUUU --------......((((((((((((((........)))))))-))..))))).............. ( -10.80, z-score = -3.34, R) >droSec1.super_0 4338635 58 + 21120651 --------CAAAUCUACCAUGAAUUAUUCCAGCGUAAAUAAUU-CAAGUGGUAAUUCGUAUUUUUUC --------......((((((((((((((........)))))))-))..))))).............. ( -10.80, z-score = -3.46, R) >droYak2.chr3R 9157840 59 - 28832112 ------AAAAAAUCUUCCAUGAAUUAUUCCACCGUAAAUAAUU-CAAGUGGUAAUUCGUAUUUUUU- ------............((((((((..((((...........-...)))))))))))).......- ( -8.34, z-score = -2.36, R) >droEre2.scaffold_4770 1250115 56 + 17746568 --------AAAAUCUACCAUGAAUUAUUCCAGCGUAAAUAAUU-CAAGUGGUAAUUCGAAUUUUU-- --------......((((((((((((((........)))))))-))..)))))............-- ( -10.80, z-score = -3.18, R) >dp4.chr2 26559263 65 + 30794189 AGUACUCGUACAUUUUCGA-GAAUUAUUCCAGCGUAAAUAAUU-CUAGAGGUAAUUCGUAUUUUUUC (((((...(((....((.(-((((((((........)))))))-)).)).)))....)))))..... ( -10.00, z-score = -1.27, R) >droPer1.super_7 119322 65 - 4445127 AGUACUCGUACAUUUUCGA-GAAUUAUUCCAGCGUAAAUAAUU-CUAGAGGUAAUUCGUAUUUUUUC (((((...(((....((.(-((((((((........)))))))-)).)).)))....)))))..... ( -10.00, z-score = -1.27, R) >droWil1.scaffold_181089 9261871 58 + 12369635 --------AAAAUUUCUUAUAAAUUACUUUAGCGUAAAUAAUU-CAAGUGGCAAUUUGGUUUCCCUU --------...............((((......)))).....(-(((((....))))))........ ( -3.60, z-score = 0.78, R) >droGri2.scaffold_14624 3809815 66 - 4233967 -AAUAAAUAACAUCUGUGUGAUUUCACACACACAUAUGCACUUACAAAUGGAUGUGUGUAUUUGUGU -.(((((((.....((((((....))))))(((((((.((........)).)))))))))))))).. ( -18.90, z-score = -1.94, R) >consensus ________AAAAUCUACCAUGAAUUAUUCCAGCGUAAAUAAUU_CAAGUGGUAAUUCGUAUUUUUUU ....................(((((((..(.................)..))))))).......... ( -3.07 = -2.73 + -0.34)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:00:29 2011